FIGURE 3.
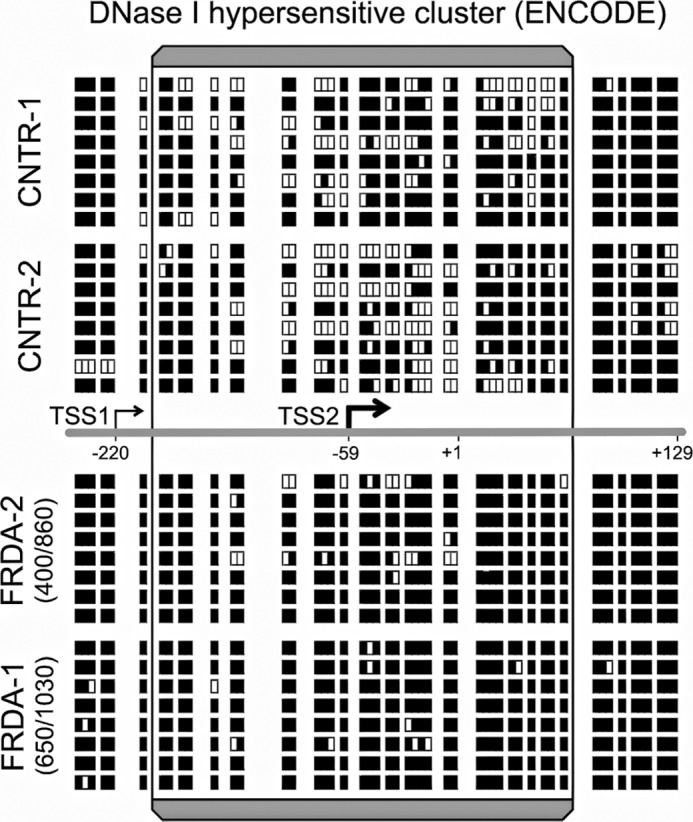
Altered nucleosome positioning in the vicinity of the major FXN-TSS and obliteration of the naturally occurring NDR in FRDA. NOMe-Seq data, measuring the nucleosomal footprint in vivo, for two non-FRDA (CNTR) and two FRDA lymphoblast cell lines are shown above and below the line depicting the region in the vicinity of FXN-TSS2 (the major FXN-TSS). The boxed region corresponds to the DNase I-hypersensitive cluster or NDR per ENCODE, within which TSS2 maps, consistent with its location in a region of relatively open chromatin configuration. NOMe-Seq data from eight clones each for the four cell lines analyzed is depicted in the form of white and black boxes, each of which represents accessible and inaccessible GpC sites, respectively (see “Experimental Procedures”). The region around TSS2 in the two non-FRDA cell lines shows many accessible GpC sites, indicating that it is in a NDR. In both FRDA cell lines, the region around TSS2 is largely inaccessible, indicating nucleosomal repositioning and obliteration of the naturally occurring NDR. Interestingly, TSS1, which maps just outside the NDR, is equally inaccessible in non-FRDA and FRDA cell lines.
