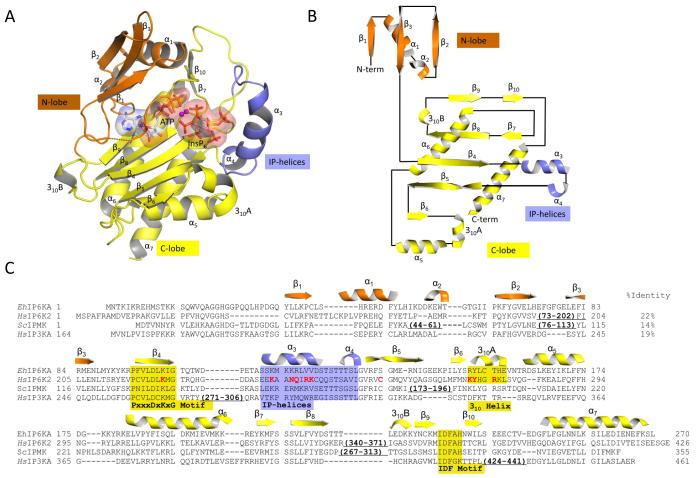
Fig. 2. Overall structure of EhIP6KA.
A, Ribbon-plot of EhIP6KA structure. ATP and InsP6 are shown as sticks within a transparent surface. Two Mg atoms are depicted as magenta balls. B, Topology diagram. C, A manual alignment of amino-acid sequences (EhIP6KA, XP-648490.2; HsIP6K2, NP_057375.2; ScIPMK, NP_010458.3; HsIP3KA, NP_002211.1), guided by the structural elements that have been observed in crystal structures, and in the case of HsIP6K2, secondary structural predictions. Outside of the conserved catalytic core of the HsIP6K2 are two significant insertions that are omitted from the alignment. The first of these (residues 73-202) includes a specialized HSP90-binding domain 54. The second insertion (residues 340-371) includes a CK2-regulated, ubiquitination motif 55. The secondary structural elements from EhIP6KA are depicted above its sequence and are color-coded orange for the N-lobe, yellow for the C-lobe and blue for the IP-helix. Structural elements that directly participate in substrate interactions are highlighted by shading. Residues in HsIP6K2 that were selected for mutagenesis are colored red. PDB codes for EhIP6KA are 4O4B, 4O4C, 4O4D, 4O4E, 4O4F.