Abstract
Polygalacturonase inhibiting proteins (PGIPs) are members of the leucine rich repeat family of proteins, involved in plant defense against fungal pathogens. PGIPs exhibit a remarkable degree of specificity in terms of their ability to bind and inhibit their target molecules, the endopolygalacturonases (EPGs). This specificity has been attributed for certain EPG/PGIP combinations to differences in primary sequence, but this explanation is unable to account for the full range of binding and inhibitory activities observed. In this paper we have fully characterized the glycosylation on the PGIP derived from Pyrus communis and demonstrated, using a combination of PNGaseF and PNGaseA in 18O-water, that the Pyrus communis PGIP utilizes all seven potential sites of N-linked glycosylation. Further, we demonstrate that certain sites appear to be modified only by glycans bearing α3-linked core fucosylation, while others are occupied by a mixture of fucosylated and non-fucosylated glycans. Modeling of the carbohydrates onto a homologous structure of PGIP indicates potential roles for glycosylation in mediating the interactions of PGIPs with EPGs.
INTRODUCTION
The plant cell wall is a major barrier against attempted invasion by phytopathogenic fungi. Therefore, the plant cell wall-degrading enzymes produced by fungi play an important role in their pathogenicity 1. Many fungi use endopolygalacturonases (EPGs) to hydrolyze the cell wall polysaccharide homogalacturonan as one of the first steps in invasion 2. A variety of plant defense mechanisms have evolved, some of which are directed toward EPGs. During pathogenesis, interactions between fungal EPGs and plant-derived polygalacturonase-inhibiting proteins (PGIPs) alter the hydrolytic activity of the EPG 3. PGIPs form high-affinity complexes with EPGs in a reversible, stoichiometric manner 3. The rate of hydrolysis of homogalacturonan by an EPG/PGIP complex is generally between one and two orders of magnitude slower than by the free EPG, depending on the source of the EPG and PGIP 4.
EPGs from a single strain of fungus may exist in a variety of isoforms 4–6. The EPG isoforms may each exist as a series of glycoforms, and may vary in their mode of action as well as in their ability to interact with, and be inhibited by, PGIPs 4, 7. For example, the EPGs from two Fusarium moniliforme isolates, though 91.7% identical, were completely different in their susceptibility to inhibition by specific PGIPs 8.
PGIPs exhibit specificity with respect to the EPGs that they bind to in vitro 4, 7, 9. The PGIPs of a single species may be present as a set of isoforms, each of which exists as a series of glycoforms 10, 11. Protein glycosylation has proven to be important in maintaining protein structure and function and can play a key role in protein-protein interactions 12, 13. This structural variability provides the potential for a wide range of specificity in EPG-PGIP interactions within any plant–pathogen pairing. The mode of action of a particular fungal EPG and its inhibition by PGIPs may be one of the critical factors in determining fungal pathogenicity. To fully understand the interactions of these two classes of molecules and their role in host-pathogen interactions the mechanisms of EPG hydrolysis of homogalacturonan and of PGIP inhibition of EPG must be understood at the molecular level.
A crystal structure of a bean PGIP has been published 14. This is an excellent starting point to begin to locate critical points of contact on the surfaces of the two proteins within the enzyme-inhibitor (EPG-PGIP) complex. We previously identified nine amino acids in the PGIPs and nine amino acids in the EPGs that are likely candidates for change due to selection pressure, thus potentially altering the specificity of interaction of the two proteins 15. This study supported data from site-specific mutation experiments 16, and indicated other regions on the PGIP that may be of importance for EPG-PGIP interaction 17. Work by others has indicated that single amino acid replacements in PGIP can alter specific EPG-PGIP interaction 16, 18, but the relevance of these findings for other EPG-PGIP combinations is currently unclear 8. Further, only a single study has investigated the role of N-linked glycosylation in EPG-PGIP binding 10 and the functional role of O-linked glycosylation in EPG-PGIP interactions has not been addressed. To date, an undisputed model of the EPG-PGIP complex has not yet been proposed 18–20. Mass spectrometry utilizing amide deuterium exchange, differential proteolysis, and modeling studies allowed us to begin to describe the structure of an EPG-PGIP complex 19.
In this paper we expand the parameters for understanding the molecular nature of the EGP-PGIP interaction by assigning N-linked glycosylation site utilization and by characterizing glycan heterogeneity at defined sites on PGIP.. This study looks at seven predicted sites of N-glycosylation found on the PGIP from Pyrus communis (pear), and demonstrates that all seven consensus sequences are utilized but that certain sites only appear to be modified by 3-linked fucosylated core glycan structures. Modeling of the carbohydrates onto a homology structure of PGIP elucidates potential roles for glycosylation in mediating the interactions of PGIPs with EPGs.
EXPERIMENTAL PROCEDURES
Preparation of PGIP peptides for N-glycosylation site mapping
PGIP from pears (Pyrus communis, cv Bartlett) was isolated and purified as described in past accounts 21. The purified protein (~ 10 µg) was resuspended in 40 mM NH4HCO3 (0.1 Ig/IL of PGIP), denatured with 1 M urea, reduced with 10 mM of DTT for 1 h at 56 °C, carboxyamidomethylated with 55 mM of iodoacetamide (Sigma) in the dark for 45 min, and then digested with 2 µg of sequencing-grade trypsin (Promega) in 40 mM NH4HCO3 overnight at 37 °C. After digestion, the peptides were acidified with 1% trifluoroacetic acid, desalted via C18 spin columns (Vydac Silica C18, The Nest Group, Inc.), and the resulting peptides were divided into two aliquots and dried down in a Speed Vac. Resulting dried peptides were resuspended in 10 µl sodium phosphate for PNGaseF or 10 µl citrate buffer for PNGaseA and then dried back down. For N-linked glycosylation site analysis, peptides were resuspended in 9 µL of 18O water (H218O, 95%, Cambridge Isotope Laboratories, Inc.) and 1 µL of N-glycanase (PNGaseF, Prozyme) and allowed to incubate for 18 hrs at 37 °C. To map the fucosylated N-linked glycosylation sites, peptides were resuspended in 9 µL of 18O water and 1 µL of N-glycanase (PNGaseA, Calbiochem) and allowed to incubate for 18 hrs at 37 °C. In either case, peptides were dried back down and resuspended in 50 µL of 40 mM of NH4HCO3, with 1 µg of trypsin, to remove any possible C-terminal incorporation of 18O from residual trypsin activity for 4 hrs 22 and then dried down and stored at −20 °C until analyzed.
Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) Analysis
The peptides were resuspended with 19.5 µL of mobile phase A (0.1% formic acid, FA, in water) and 0.5 µL of mobile phase B (80% acetonitrile, ACN, and 0.1% FA in water) and filtered with 0.2 µm filters (Nanosep, PALL). Proteins were analyzed as previously described 23. Briefly, the samples were loaded off-line onto a nanospray tapered capillary column/emitter (360 × 75 × 15 µm, PicoFrit, New Objective) self-packed with C18 reverse-phase (RP) resin (8.5 cm, Waters) in a Nitrogen pressure bomb for 10 min at 1,000 psi (~5 µL load) and then separated via a 160 min linear gradient of increasing mobile phase B at a flow rate of ~200 nL/min directly into the mass spectrometer. One-dimensional LC-MS/MS analysis was performed on a linear ion trap mass spectrometer (LTQ, Thermo Fisher Scientific Inc., San Jose, CA) equipped with a nanoelectrospray ion source. A full MS spectrum was collected (m/z 350–2000) followed by 8 MS/MS spectra following CID (34% normalized collision energy) of the most intense peaks. Dynamic exclusion was set at 2 for 30 s exclusion.
Data Analysis
The resulting data was searched against a database containing the polygalacturonase-inhibiting protein (PGIP, gi|33087512 and Pyrus communis) fasta sequence, obtained from NCBI, and the common contaminants database (ThermoFisher) using the TurboSequest algorithm (BioWorks 3.1, Thermo Fisher Scientific Inc.). Spectra with a threshold of 15 ions and a TIC of 2e3 were search. Charge state analysis using ZSA, correct ion, combion, and ionquest (all as part of the Bioworks work flow) were applied over a range of [MH]+ = 600–4000. The SEQUEST parameters were set to allow 2.0 Da of precursor ion mass tolerance and 0.5 Da of fragment ion tolerance with monoisotopic mass. Only fully-tryptic peptides were allowed with up to three missed internal cleavage sites. Dynamic mass increases of 15.99 and 57.02 Da were allowed for oxidized methionine and alkylated cysteine, respectively. Carbamylation, due to the presence of urea, was scanned for but not observed. In the cases where sites of N-linked glycosylation were investigated with N-glycanase and 18O water, a dynamic mass increase of 3.00 Da was allowed for Asn residues as previously described 24. The results of the SEQUEST search were filtered at 2.0/2.3/2.8 Xcorr (Cross correlation) with charge states of +1/+2/+3, respectively. Only Asn residues in consensus sites were identified as being modified by +3 Da and all peptide fragmentation data indicative of a site of modification was manually validated following automated filtering. All spectra leading to assignments are available upon request.
N-Glycan Release, Permethylation, and Analysis
Glycans were released from PGIP by sequential enzymatic digestion, first with PNGaseF (Prozyme) and subsequently with PNGaseA (Calbiochem). Preparation and analysis of N-linked glycans was performed as previously described 25. Briefly, between 5 and 10 µg of PGIP were denatured by boiling and incubated with trypsin. The resulting digest was subjected to reverse phase chromatography to enrich for glycopeptides, which were then concentrated to dryness by vacuum centrifugation before treatment with PNGaseF. Exhaustive digestion with PNGaseF was performed in 20 mM sodium phosphate, pH 7.5. The reaction mixture was subjected to reverse phase chromatography to separate the PNGaseF-sensitive glycans from residual glycopeptide and from deglycosylated peptide. The residual glycopeptide/peptide mixture was then digested with PNGaseA in 0.2M citrate phosphate buffer, pH 5.0. Released glycans were separated from residual peptide by reverse phase chromatography, yielding PNGaseA-sensitive glycans. Both pools of glycan were permethylated and analyzed by nanospray ionization tandem mass spectrometry, NSI-MSn, using a linear ion trap mass spectrometer (LTQ, ThermoFisher). All spectra leading to assignments are available upon request.
Homology Modeling
To create a homology model for PGIP the mature amino acid sequence of pear PGIP was threaded onto the crystal structure of PGIP from P. vulgaris 14 using the Swiss PDB Viewer, DeepView v. 3.7. The homology structure was then optimized with molecular mechanics, MM3, using the protein modeling suite, BioMedCache v. 6.1 (BMC). The optimized structure was then allowed to move using molecular dynamics, simulating 300 K for 2 ps, before it was again optimized with molecular mechanics. Seven glycosylation, three M3XN2 (Man3XylGlcNAc2) and four M3XN2F (Man3XylGlcNAc2Fuc), were created using BMC, and their structures were optimized using the semi-empirical method, PM5. The N-linked glycosylations were attached to the appropriate residues and the entire glycosylated PGIP model was optimized by applying several iterations of molecular mechanics and molecular dynamics computations as before.
RESULTS
N-linked Site Mapping of PGIP with PNGAseF
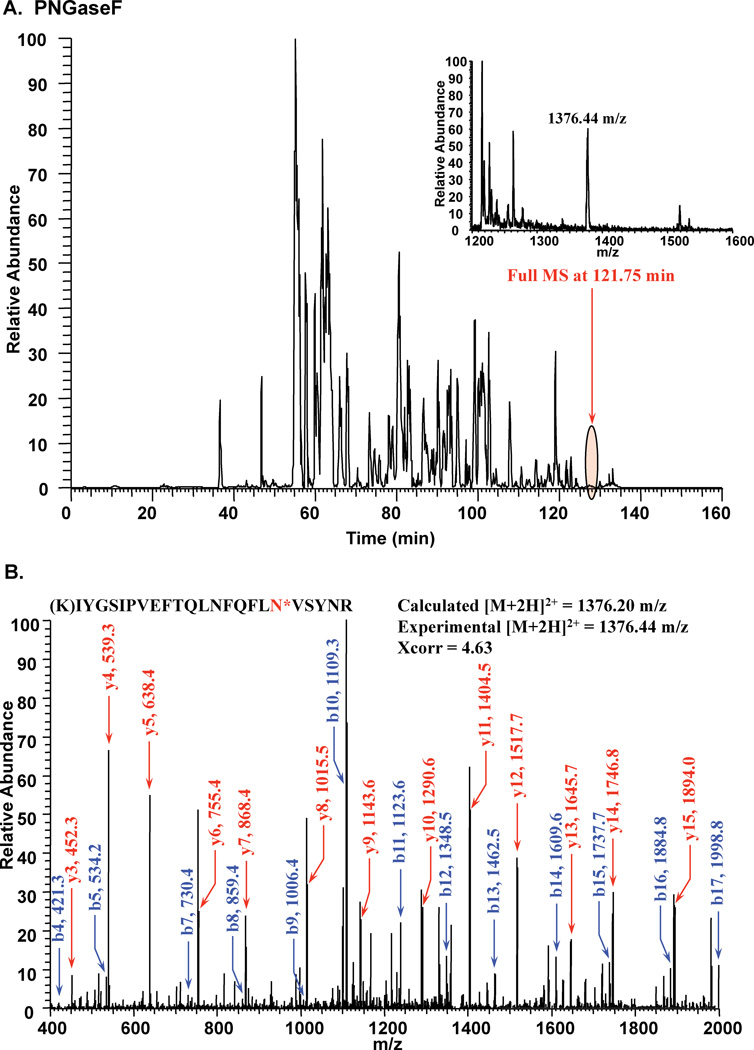
Pyrus communis PGIP has seven potential N-glycoslyation sites (N-X-S/T, where X is not P). In order to determine whether consensus sites were being utilized, purified, digested PGIP-derived peptides were treated with PNGaseF in 18O-water to convert modified Asn residues to heavy Asp (a mass shift of 3 Da). Following enzymatic treatment, peptides were analyzed by LC-MS/MS and 3 of the potential 7 Asn were determined to be sites of N-linked glycosylation (Fig. 1, Table 1). However, it was noted that complete coverage of the protein was only 61% and that tryptic peptides containing the other 4 potential sites of modification were not identified in this experiment (Table 1).
Figure 1.
Tandem mass spectrometry following PNGase F treatment in 18O-water to map sites of N-linked glycosylation on PGIP. A. Full chromatogram of LC-MS/MS run. B. MS/MS fragmentation leading to the identification of a site of modification (N*= +3 Da).
Table 1.
Sites of N-linked Glycosylation and Sequence Coverage of PGIP
 |
N-linked Site Mapping of PGIP with PNGaseA
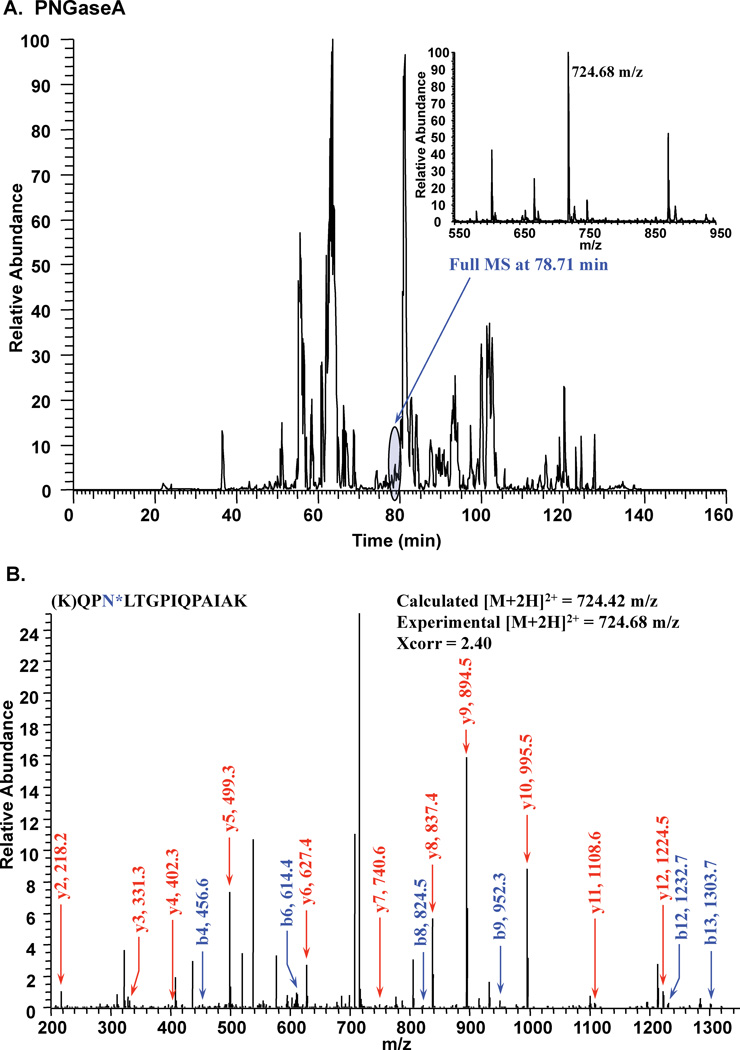
It is well established that both plants and insects, unlike mammals, are capable of α3-linked fucose modification of the reducing terminal GlcNAc residue in N-linked structures 26. Furthermore, PNGaseF is unable to cleave N-linked glycans containing α 3-linked fucose in the chiotbiose core 27. Therefore, we utilized PNGaseA to incorporate 18O at glycosylated Asn sites. This enzyme is able to cleave 3-linked fucose on the reducing end GlcNAc of N-linked structures 27. Following PNGaseA treatment in 18O-water of the PGIP-derived peptides, coverage increased to 81% and all 7 sites of N-linked glycosylation were shown to be utilized (Fig. 2, Table 1).
Figure 2.
Tandem mass spectrometry following PNGase A treatment in 18O-water to map sites of N-linked glycosylation on PGIP. A. Full chromatogram of LC-MS/MS run. B. MS/MS fragmentation leading to the identification of a site of modification (N*= +3 Da).
Glycan Analysis following PNGaseF and PNGaseA release from PGIP
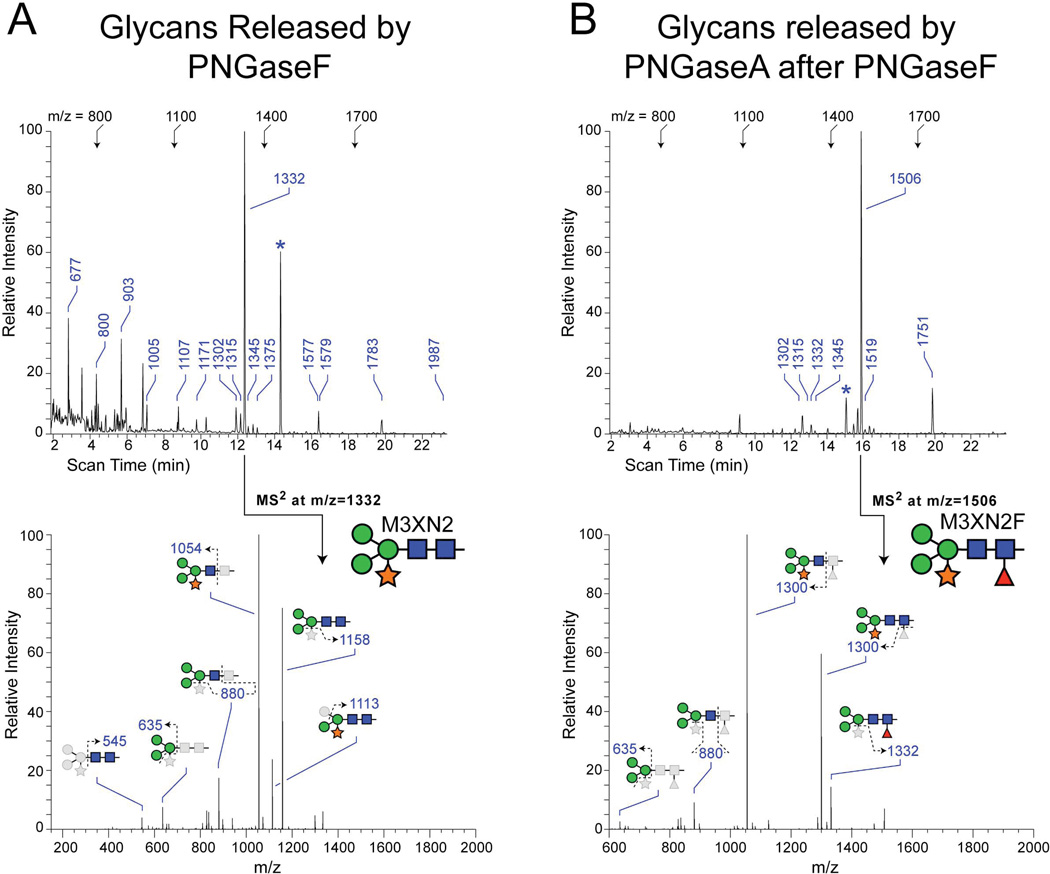
In order to determine the full range of structures that might be found at the 7 N-linked glycosylation sites of PGIP, glycans were released by PNGaseF or by PNGaseA (following PNGaseF). Released, permethylated glycans were analyzed by tandem mass spectrometry 25. The profile of glycans released from PGIP by PNGaseF digestion is dominated by a single structure, the xylosylated tri-mannosyl core glycan M3XN2 (Figure 3, m/z = 1331). Fragmentation analysis places the Xyl residue onto the branched Man residue, as previously described for plant N-linked glycans 28. In addition to the dominant PNGaseF-sensitive glycan, 14 other detectable glycans were released by PNGaseF digestion. A full series of high-mannose glycans, ranging from M9N2 to M3N2, as well as hybrid (NM3N2, NM3XN2) and complex (N2M3N2) glycans were released by PNGaseF, but were detected at very low levels (Table 2). Following exhaustive treatment with PNGaseF, digestion of residual glycoprotein with PNGaseA identified a single dominant PNGaseF-resistant, PNGaseA-sensitive glycan as M3XN2F, where the Fuc residue was detected in α3-linkage to the reducing terminal GlcNAc residue (Figure 2B, m/z = 1506). The remaining detectable, but minor, PNGaseA-sensitive glycans all share a characteristic α3-linked Fuc on the reducing terminal core GlcNAc residue (Table 2). Both paucimannose (M3N2, M2N2) and hybrid-type (NM3XN2) glycans are modified with core Fucose on PGIP. The diversity of minor glycans released by PNGaseF or PNGaseA is consistent with the microheterogeneity inherent in N-linked glycosylation. However, the overwhelming predominance of M3XN2 and M3XN2F indicates that PGIP exhibits a significant level of glycosylation homogeneity.
Figure 3.
N-linked glycan analysis of PGIP. A. Total ion mapping (TIM) profile and MS/MS fragmentation of most abundant glycan following PNGAseF release. B. TIM profile and MS/MS fragmentation of most abundant glycan following PNGaseA release after removal of PNGaseF released glycans. For TIM scans in A and B, landmark m/z values are shown across the top at their equivalent scan times for each sample. For detected glycans, m/z values are indicated along the scan. Asterisks indicate a non-glycan contaminant at m/z=1452, which is routinely detected following permethylation. For MS/MS spectra, glycan representations are consistent with the nomenclature recommendations of the Consortium for Functional Glycomics (Man, green circle; GlcNAc, blue square; Fuc, red triangle; Xyl, orange star). The dominant glycan released by PNGaseF is M3XN2. For PNGaseA, the dominant glycan is the α3-fucosylated version of the same structure, M3XN2F.
Table 2.
N-linked glycans of PGIP
| Glycan Structure | Permethylated Mass (m/z) |
Glycan Prevalence (% Total Profile) |
||
|---|---|---|---|---|
| Singly charged [m+Na]+ |
Doubly charged [m+2Na]2+ |
Released by PNGaseF |
Released by PNGaseA after PNGaseF |
|
| 1171 | nd1 | 1.9 | nd | |
| 1375 | nd | 1.6 | nd | |
| 1579 | 800 | 5.63 | nd | |
| 1783 | 903 | 17.4 | nd | |
| 1987 | 1005 | 3.3 | nd | |
| 2191 | 1107 | 1.5 | nd | |
| 2395 | nd | <0.34 | nd | |
| 1416 | nd | <0.3 | nd | |
| 1620 | nd | <0.3 | nd | |
| 1684 | nd | <0.3 | nd | |
| 1661 | nd | <0.3 | nd | |
| 1127 | nd | 1.3 | nd | |
| 1331 | 677 | 61.8 | nd | |
| 1576 | 800 | 5.63 | nd | |
| 1141 | nd | <0.3 | <0.3 | |
| 1345 | nd | nd | 1.3 | |
| 1302 | nd | nd | 3.8 | |
| 1506 | nd | nd | 87.5 | |
| 1750 | nd | nd | 5.5 | |
| 1315 | nd | nd | 0.6 | |
| 1519 | nd | nd | 1.4 | |
”nd” denotes “not detected”
Superscripted “3” or “6” indicates the Fuc linkage position onto GlcNAc.
The M5N2 and NM3XN2 glycans were primarily detected as doubly-charged ions, which were unresolved (m/z = 800 for both). Therefore, the calculated prevalence was divided equally between the two glycans.
Glycans with prevalence indicated as “<0.3” were detected below the threshold of quantification, which was taken as 2 times background.
DISCUSSION
The differences in the specificities of EPGs for PGIPs have generally been attributed to changes in individual amino acids within individual PGIPs 29 and EPGs 30. These single site changes are unlikely to account for the wide spectrum of binding and inhibitory activities observed, and little emphasis has been placed on the potential for glycosylation to effect the ability of PGIPs to bind, and/or to inhibit EPGs. We have previously discovered that several of the EPGs we have studied (PG I, PGA, PGC from A. niger, along with PG 2 and PG 3 from B. cinerea) contain O-linked mannose 31–33. These modifications are located near the N-terminus in close proximity to the sites where we hypothesize PGIP binding occurs. This has led us to speculate that O-mannosylation may play an important role in EPG/PGIP interaction, as the O-Man-initiated modification has been previously observed to have a dramatic impact on the α-dystroglycan/laminin interaction in animals 13.
An earlier study provided partial characterization of the carbohydrate structures present on two N-linked sites in Phaseolus vulgaris (bean) PGIP by MALDI-TOF MS 34. Here we have demonstrated that the Pyrus communis PGIP utilizes all seven sites of glycosylation by using a combination of PNGaseF and PNGaseA to sequentially release differentially sensitive glycans. By digesting PGIP with PNGaseF or A in 18O-water modified Asn residues are converted to 18O-Asp, tagging the site of modifications with a mass label (+3 Da, Figs. 1–2, Table 1). Our methodology clearly illustrates the need to use the less commonly used N-glycanase, PNGaseA, to fully site-map and characterize the N-linked glycans in plants (and insects). Interestingly, 4 of the 7 sites appear to be exclusively modified by fucosylated N-liked glycans as they were not detected following PNGaseF treatment. While the major PNGAseF sensitive glycan was M3XN2 and the major PNGaseA sensitive glycan was M3XN2F (with the fucose in α 3-linkage), several other minor glycan structures, including those observed in the earlier study on Phaseolus vulgaris PGIP were observed (Table 2).
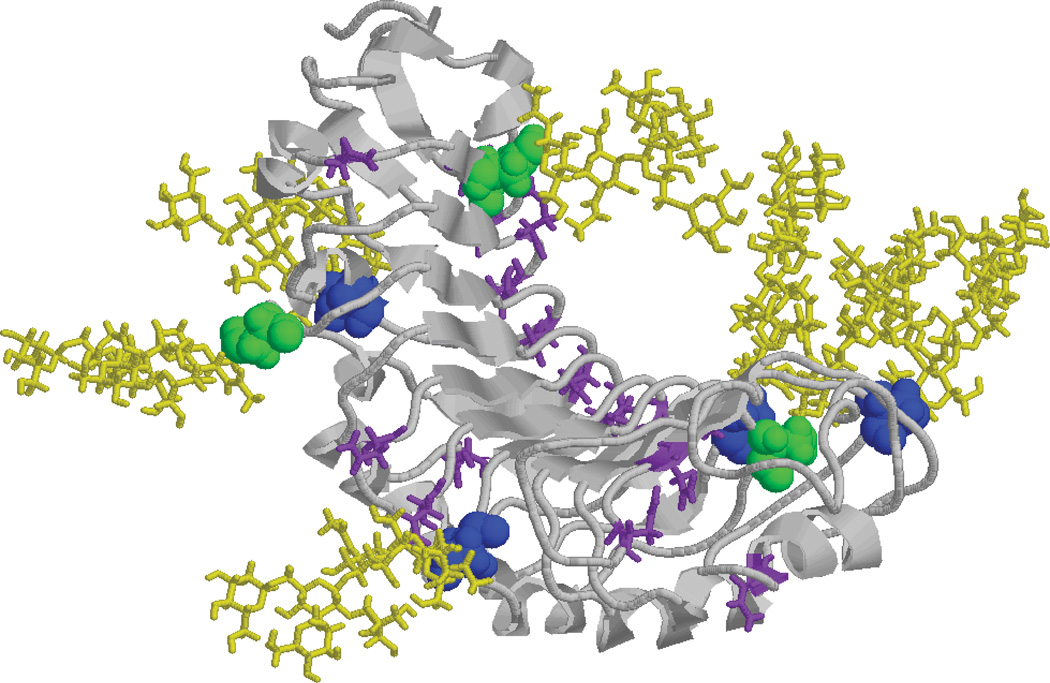
A homology model with the most abundant glycan structures was generated and energy minimized to better understand the impact of N-linked glycosylation on the interaction interface (Fig. 4). Inspection of the model generated indicates that the N-linked glycans of Pyrus communis PGIP lie on either side of the binding surface, greatly increasing the surface area available for interaction with the target EPG, but also increasing the potential for steric modulation of binding by glycosylation. The striking dominance of α3-fucosylated glycans at specific N-linked sites suggests that this Fuc residue imparts protein conformation that is favored for interactions with EPG. Extremely little is known about the factors that influence whether individual N-linked glycans are modified by core Fuc. Elucidation of PGIP glycosylation lays a groundwork for investigating whether specific protein motifs favor, or perhaps inhibit, the elaboration of α3-Fuc.
Figure 4.
Homology Model of N-glycosylated PGIP. PNGaseA-only sensitive sites are shown in blue with M3XN2F attached in yellow and PNGAseF and A sensitive sites are shown in green with M3XN2 attached in yellow. The peptide backbone is shown in grey with all unmodified Asn residues labeled in purple.
Pyrus communis PGIP is relatively heavily glycosylated, yet it also appears to be designed to allow for additional glycosylation. The asparagines running down either side of the molecule are components of what is termed an asparagine ladder which is believed to add stability to the structure 14. Not all of these asparagines fall into an N-linked glycosylation consensus sequence (NXS/T), but each asparagine provides a site for evolution toward the generation of a new glycosylation site. The situation is similar for the Phaseolus vulgaris PGIP which also displays an asparagine ladder 14, and PGIPs are known to exist in multiple glycosylation states 14, 15. Whether glycosylation directly impacts the ability to bind and /or inhibit EPGs awaits further analysis, but the significant presence of specific N-linked glycans flanking the PGIP/EPG interaction region requires that their contribution be considered in elucidating the biological specificity of this host-pathogen interaction.
ACKNOWLEDGEMENTS
We would like to thank all members of the Bergmann, Tiemeyer, and Wells laboratories as well as Ron Orlando for helpful discussions. This work was supported in part by a grant from NIH/NCRR (5P41RR018502, L.W. and M.T. senior investigators) and from NIH/NIGMS (1-R01-GM072839 to M.T.), and from the DOE (DE-FG02-96ER20221 and DE-FG02-93ER20097, C.B. co-PI).
REFERENCES
- 1.Cooper RM. Biochemical Plant Pathology. 1985. The mechanisms and signficance of enzymic degradation of host cell walls by parasites; p. 135. [Google Scholar]
- 2.Jones TM, Anderson AJ, Albersheim P. Host-Pathogen Interactions IV. Studies of the polysaccharide-degrading enzymes. Physiol. Plant Pathol. 1972;2:153–166. [Google Scholar]
- 3.Cervone F, De Lorenzo G, Aracri B, Bellincampi D, Caprari C, Devoto A, Mattei B, Nuss L, Salvi G. Biology of Plant-Microbe Interactions. 1996. The PGIP Family: Extracellular proteins specialized for recognition; pp. 93–98. [Google Scholar]
- 4.Cook BJ, Clay RP, Bergmann CW, Albersheim P, Darvill AG. Fungal polygalacturonases exhibit different substrate degradation patterns and differ in their susceptibilities to polygalacturonase-inhibiting proteins. Mol Plant Microbe Interact. 1999;12(8):703–711. doi: 10.1094/MPMI.1999.12.8.703. [DOI] [PubMed] [Google Scholar]
- 5.Centis S, Dumas B, Fournier J, Marolda M, Esquerre-Tugaye MT. Isolation and sequence analysis of Clpg1, a gene coding for an endopolygalacturonase of the phytopathogenic fungus Colletotrichum lindemuthianum. Gene. 1996;170(1):125–129. doi: 10.1016/0378-1119(95)00867-5. [DOI] [PubMed] [Google Scholar]
- 6.Parenicova L, Benen JA, Kester HC, Visser J. pgaA and pgaB encode two constitutively expressed endopolygalacturonases of Aspergillus niger. Biochem J. 2000;345 Pt 3:637–644. [PMC free article] [PubMed] [Google Scholar]
- 7.Kemp G, Stanton L, Bergmann CW, Clay RP, Albersheim P, Darvill A. Polygalacturonase-inhibiting proteins can function as activators of polygalacturonase. Mol Plant Microbe Interact. 2004;17(8):888–894. doi: 10.1094/MPMI.2004.17.8.888. [DOI] [PubMed] [Google Scholar]
- 8.Sella L, Castiglioni C, Roberti S, D'Ovidio R, Favaron F. An endo-polygalacturonase (PG) of Fusarium moniliforme escaping inhibition by plant polygalacturonase-inhibiting proteins (PGIPs) provides new insights into the PG-PGIP interaction. FEMS Microbiol Lett. 2004;240(1):117–124. doi: 10.1016/j.femsle.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Kemp G, Bergmann CW, Clay R, Van der Westhuizen AJ, Pretorius ZA. Isolation of a polygalacturonase-inhibiting protein (PGIP) from wheat. Mol Plant Microbe Interact. 2003;16(11):955–661. doi: 10.1094/MPMI.2003.16.11.955. [DOI] [PubMed] [Google Scholar]
- 10.Bergmann C, Cook BJ, Darvill A, Albersheim P, Bellincampi D, Caprari C. The effect of glycosylation of EPG and PGIP on the production of oligogalacturonides. Pectins and Pectinases. 1996:275–282. [Google Scholar]
- 11.Desiderio A, Aracri B, Leckie F, Mattei B, Salvi G, Tigelaar H, Van Roekel JS, Baulcombe DC, Melchers LS, De Lorenzo G, Cervone F. Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol Plant Microbe Interact. 1997;10(7):852–860. doi: 10.1094/MPMI.1997.10.7.852. [DOI] [PubMed] [Google Scholar]
- 12.Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 13.Martin PT. Dystroglycan glycosylation and its role in matrix binding in skeletal muscle. Glycobiology. 2003;13(8):55R–66R. doi: 10.1093/glycob/cwg076. [DOI] [PubMed] [Google Scholar]
- 14.Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, De Lorenzo G, Tsernoglou D, Cervone F. The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci U S A. 2003;100(17):10124–10128. doi: 10.1073/pnas.1733690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stotz HU, Bishop JG, Bergmann C, Koch M, Albersheim P, Darvill A, Labavitch JM. Identification of target amino acids that affect interactions of fungal polygalacturonases and their plant inhibitors. Physiol. Mol. Plant Pathol. 1999;56:117–130. [Google Scholar]
- 16.Leckie F, Mattei B, Capodicasa C, Hemmings A, Nuss L, Aracri B, De Lorenzo G, Cervone F. The specificity of polygalacturonase-inhibiting protein (PGIP): a single amino acid substitution in the solvent-exposed beta-strand/beta-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. Embo J. 1999;18(9):2352–2363. doi: 10.1093/emboj/18.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop JG. Directed mutagenesis confirms the functional importance of positively selected sites in polygalacturonase inhibitor protein. Mol Biol Evol. 2005;22(7):1531–1534. doi: 10.1093/molbev/msi146. [DOI] [PubMed] [Google Scholar]
- 18.Federici L, Caprari C, Mattei B, Savino C, Di Matteo A, De Lorenzo G, Cervone F, Tsernoglou D. Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase-inhibiting protein) Proc Natl Acad Sci U S A. 2001;98(23):13425–13430. doi: 10.1073/pnas.231473698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King D, Bergmann C, Orlando R, Benen JA, Kester HC, Visser J. Use of amide exchange mass spectrometry to study conformational changes within the endopolygalacturonase II-homogalacturonan-polygalacturonase inhibiting protein system. Biochemistry. 2002;41(32):10225–10233. doi: 10.1021/bi020119f. [DOI] [PubMed] [Google Scholar]
- 20.Sicilia F, Fernandez-Recio J, Caprari C, De Lorenzo G, Tsernoglou D, Cervone F, Federici L. The polygalacturonase-inhibiting protein PGIP2 of Phaseolus vulgaris has evolved a mixed mode of inhibition of endopolygalacturonase PG1 of Botrytis cinerea. Plant Physiol. 2005;139(3):1380–1388. doi: 10.1104/pp.105.067546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stotz HU, Powell AL, Damon SE, Greve LC, Bennett AB, Labavitch JM. Molecular characterization of a polygalacturonase inhibitor from Pyrus communis L cv Bartlett. Plant Physiol. 1993;102(1):133–138. doi: 10.1104/pp.102.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angel PM, Lim JM, Wells L, Bergmann C, Orlando R. A potential pitfall in 18O-based N-linked glycosylation site mapping. Rapid Commun Mass Spectrom. 2007;21(5):674–682. doi: 10.1002/rcm.2874. [DOI] [PubMed] [Google Scholar]
- 23.Lim JM, Sherling D, Teo CF, Hausman DB, Lin D, Wells L. Defining the regulated secreted proteome of rodent adipocytes upon the induction of insulin resistance. J Proteome Res. 2008;7(3):1251–1263. doi: 10.1021/pr7006945. [DOI] [PubMed] [Google Scholar]
- 24.Koles K, Lim JM, Aoki K, Porterfield M, Tiemeyer M, Wells L, Panin V. Identification of N-glycosylated proteins from the central nervous system of Drosophila melanogaster. Glycobiology. 2007;17(12):1388–1403. doi: 10.1093/glycob/cwm097. [DOI] [PubMed] [Google Scholar]
- 25.Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282(12):9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 26.Tomiya N, Narang S, Lee YC, Betenbaugh MJ. Comparing N-glycan processing in mammalian cell lines to native and engineered lepidopteran insect cell lines. Glycoconj J. 2004;21(6):343–360. doi: 10.1023/B:GLYC.0000046275.28315.87. [DOI] [PubMed] [Google Scholar]
- 27.Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180(2):195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 28.Kurosaka A, Yano A, Itoh N, Kuroda Y, Nakagawa T, Kawasaki T. The structure of a neural specific carbohydrate epitope of horseradish peroxidase recognized by anti-horseradish peroxidase antiserum. J Biol Chem. 1991;266(7):4168–4172. [PubMed] [Google Scholar]
- 29.Di Matteo A, Bonivento D, Tsernoglou D, Federici L, Cervone F. Polygalacturonase-inhibiting protein (PGIP) in plant defence: a structural view. Phytochemistry. 2006;67(6):528–533. doi: 10.1016/j.phytochem.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 30.Raiola A, Sella L, Castiglioni C, Balmas V, Favaron F. A single amino acid substitution in highly similar endo-PGs from Fusarium verticillioides and related Fusarium species affects PGIP inhibition. Fungal Genet Biol. 2008;45(5):776–789. doi: 10.1016/j.fgb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Colangelo J, Licon V, Benen J, Visser J, Bergmann C, Orlando R. Characterization of the glycosylation of recombinant endopolygalacturonase I from Aspergillus niger. Rapid Commun Mass Spectrom. 1999;13(14):1448–1553. doi: 10.1002/(SICI)1097-0231(19990730)13:14<1448::AID-RCM665>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 32.Woosley B, Xie M, Wells L, Orlando R, Garrison D, King D, Bergmann C. Comprehensive glycan analysis of recombinant Aspergillus niger endo-polygalacturonase C. Anal Biochem. 2006;354(1):43–53. doi: 10.1016/j.ab.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Woosley BD, Kim YH, Kumar Kolli VS, Wells L, King D, Poe R, Orlando R, Bergmann C. Glycan analysis of recombinant Aspergillus niger endo-polygalacturonase A. Carbohydr Res. 2006;341(14):2370–2378. doi: 10.1016/j.carres.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Mattei B, Bernalda MS, Federici L, Roepstorff P, Cervone F, Boffi A. Secondary structure and post-translational modifications of the leucine-rich repeat protein PGIP (polygalacturonase-inhibiting protein) from Phaseolus vulgaris. Biochemistry. 2001;40(2):569–576. doi: 10.1021/bi0017632. [DOI] [PubMed] [Google Scholar]