Abstract
The tumour suppressor EWI2 associates with tetraspanins and regulates tumour cell movement and proliferation. The short cytoplasmic domain of EWI2 is positively charged; five out of the ten residues of this domain are basic. In the present study we demonstrated that the EWI2 cytoplasmic tail interacts specifically with negatively charged PIPs (phosphatidylinositol phosphates), but not with other membrane lipids. The PIPs that interact with EWI2 cytoplasmic tail include PtdIns5P, PtdIns4P, PtdIns3P, PtdIns(3,5)P2 and PtdIns(3,4)P2. The binding affinity of PIPs to the EWI2 tail, however, is not solely based on charge because PtdIns5P, PtdIns4P and PtdIns3P have a higher affinity to EWI2 than PtdIns(3,5)P2 and PtdIns(3,4)P2 do. Mutation of either of two basic residue clusters in the EWI2 cytoplasmic tail abolishes PIP binding, and PIP binding is also determined by the position of basic residues in the EWI2 cytoplasmic tail. In addition, EWI2 is constitutively palmitoylated at the cytoplasmic cysteine residues located at the N-terminal of those basic residues. The PIP interaction is not required for, but appears to regulate, the palmitoylation, whereas palmitoylation is neither required for nor regulates the PIP interaction. Functionally, the PIP interaction regulates the stability of EWI2 proteins, whereas palmitoylation is needed for tetraspanin–EWI2 association and EWI2-dependent inhibition of cell migration and lamellipodia formation. For cell–cell adhesion and cell proliferation, the PIP interaction functions in opposition to the palmitoylation. In conclusion, the EWI2 cytoplasmic tail actively engages with the cell membrane via PIP binding and palmitoylation, which play differential roles in EWI2 functions.
Keywords: immunoglobulin superfamily (IgSF), palmitoylation, phospholipids, tetraspanin
INTRODUCTION
EWI2 or CD316 is a newly identified member of the IgSF (immunoglobulin superfamily) and physically associates with tetraspanins such as CD9, CD81 and CD82 [1–4]. EWI2 shares a relatively high amino acid sequence homology with other IgSF proteins including FPRP/CD9P-1, IgSF3 and CD101, and thus forms a novel Ig subfamily called the EWI Ig subfamily with these proteins [1–4]. Among EWI subfamily members, FPRP/CD9P-1 was also found to associate with tetraspanins [5,6]. Whether or not the rest of the members in the EWI subfamily, such as IgSF3 and CD101, bind to tetraspanins remains to be tested. Although the biological functions of EWI proteins are not understood, the associations with tetraspanins indicate that EWI proteins might influence tetraspanin functions such as cell fusion, adhesion and migration [7–11]. For instance, EWI2 overexpression inhibits tumour cell migration on LN (laminin), fibronectin and collagen, whereas silencing of EWI2 promotes tumour-cell migration [3,12,13]. Because regulating cell migration is a prominent feature of tetraspanins, EWI2 may regulate motility through them. However, as an IgSF protein, EWI2 itself may function as a cell–cell adhesion molecule and be involved in the interaction between cells in organs such as the brain, kidney and testis, in all of which EWI2 is highly expressed [3]. In fact, a previous study demonstrated that heat-shock protein A8, which can be expressed as a cell-bound form, serves as a ligand for EWI2 at the surface of dendritic cells [14]. Moreover, the expression of EWI2 is down-regulated upon malignant transformation in the brain and is associated with better prognosis for patients with brain tumours [15]. Forced expression of EWI2 inhibits brain tumour growth in an animal model [15].
Although EWI2 and tetraspanin associations are well documented, the structural elements that determine the physical interactions between EWI2 and tetraspanins remain controversial [1,2]. The ectodomain of EWI2 [1], the cytoplasmic tail of EWI2 [12,16] and the transmembrane regions of tetraspanins [4] have been reported to be either critical or important for EWI2–tetraspanin associations. It remains to be determined the reasons that EWI2–tetraspanin associations can be disrupted by 1 %Triton X-100 or 1 %Nonidet P40 detergent extraction [1–4] if the direct protein–protein interactions are mediated by their ectodomains. Nevertheless, the short cytoplasmic domain of EWI2 has clear functional impact. Swapping the EWI2 tail with the CD2 cytoplasmic domain results in the reversal of the phenotypes of wild-type EWI2, such as the inhibition of cell migration and cell spreading, and the association with tetraspanin and integrin [12,16]. Distinctive from other EWI subfamily members, EWI2 contains a 10-amino acid short cytoplasmic domain. Of the ten residues, five are positively charged residues, i.e. lysine and arginine, and two are cysteine residues that are located, presumably, immediately proximal to the inner leaflet of the membrane. These residues are fully conserved between murine and human EWI2 proteins. Because the inner leaflet of the membrane contains a variety of negatively charged acidic lipids such as PIPs (phosphatidylinositol phosphates), it becomes important to determine whether or not the positively charged EWI2 cytoplasmic domain interacts directly with the negatively charged lipids in the membrane. Meanwhile, based on the knowledge that the membrane-proximal cytoplasmic cysteine residues can be palmitoylated, we predict that EWI2 undergoes palmitoylation at the cysteine residues of its cytoplasmic domain. Indeed, both hypotheses proved correct. In addition, we addressed the mutual regulation and biological functions of the acidic lipid interaction and palmitoylation of EWI2.
MATERIALS AND METHODS
Cells and transfectants
The NIH 3T3 cell line was obtained from A.T.C.C. and cultured at 37°C and 5 % CO2 in DMEM (Dulbecco’s modified Eagle’s medium; Invitrogen) supplemented with 10 % FBS (fetal bovine serum), 100 units/ml penicillin and 100 µg/ml streptomycin. EWI2 wild-type and cytoplasmic domain mutants were constructed into a eukaryotic expression vector pZeoSV2 (Invitrogen) as described previously [17]. Briefly, EWI2 cytoplasmic mutants were generated by replacing through PCR either cysteine or positive residues in the wild-type EWI2 cytoplasmic domain with alanine residues. The full-length cDNAs encoding human wild-type and mutant EWI2 were constructed into the pZeoSV2 vector through the HindIII and XbaI sites respectively. The EWI2 coding information was confirmed by DNA sequencing. NIH 3T3 cells were transfected with plasmid DNA by using Lipofectamine™ 2000 (Invitrogen) and selected with zeocin (Invitrogen) at a concentration of 0.2 mg/ml. Hundreds of zeocin-resistant clones were pooled, and the human EWI2-positive clones were collected by flow cytometric cell sorting. The pooled and sorted EWI2-positive clones were the stable transfectants of EWI2 wild-type or mutants used in the following experiments. The GFP (green fluorescent protein) fusion constructs of EWI2 wild-type and mutants were generated by fusing the EWI2 cDNAs upstream of the GFP sequence in the pEGFP-N2 vector (BD Clontech). The stable transfectants of GFP and EWI2–GFP fusions were established in NIH 3T3 cells as described above by collecting through flowcytometric cell sorting GFP-positive clones resistant to geneticin (Invitrogen).
Antibodies, ECM (extracellular matrix) proteins and other reagents
The mAbs (monoclonal antibodies) used in the present study were human anti-EWI2 8A12 [3] and 5E8 [14] (kindly provided by Dr T. Schweighoffer of Norvatis, Basel, Switzerland), mouse anti-CD44 KM114 (BD Pharmingen), anti-PtdIns4P IgM-type (Echelon Biosciences) and a negative control mAb mouse ant-IgG2b (clone MOPC 141) (Sigma). The pAb (polyclonal antibody) against human EWI2 was generated by using the EWI2 ectodomain as the immunogen and purified by using affinity chromatography. Rhodamine- or FITC (fluorescein isothiocyanate)-conjugate goat anti-mouse IgG and IgM were purchased from Biosource International. HRP (horseradish peroxidase)-conjugated anti-mouse IgG and ExtrAvidin were from Sigma. The extracellular matrix proteins were human plasma fibronectin and mouse LN 1 (Life Technologies). Ins(4,5)P2 and Ins4P were purchased from Echelon Biosciences, hydroxylamine HCl was purchased from Pierce, and Pipes was from Sigma.
Protein-lipid overlay assay
The protein-lipid overlay assay was performed using the synthetic N-terminally biotinylated peptides containing the EWI2 cytoplasmic domain sequence and the PIP, and Sphingo strips and/or arrays (Echelon Biosciences). The peptide- and lipid-binding experiments were performed according to the manufacturer’s instructions. Briefly, the PIP and Sphingo strip and/or array membranes were blocked with TBS buffer [10 mM Tris/HCl (pH 8.0) and 150 mM NaCl] containing 3 % fatty-acid-free BSA (Sigma) and 0.1 % Tween 20 for 1 h at room temperature (22 °C). The membranes were then incubated with 0.05–0.5 µg/ml of the biotinylated peptide in the same buffer for 1 h at room temperature or overnight at 4°C. After three washes with the same buffer, the membranes were incubated with HRP-conjugated extravidin, followed by three washes. The bound peptides were detected with an ECL (enhanced chemiluminescence) kit from Amersham Pharmacia.
The labelling of the EWI2 cytoplasmic domain peptide with palmitate was performed as described previously [18] with modifications. The peptide (5.4 mg) was incubated with N-succinimidyl palmitate (15 mg) (Toronto Research Chemicals) in 2 ml of anhydrous dimethylformamide at 4°C overnight. A 950-µl aliquot of the reaction product was mixed with 950 µl of H2O and 1.9 ml of ethyl acetate, and the mixture was stilled for 1–2 min until the layers were clearly differentiated. The top layer, i.e. the ethyl acetate phase, was carefully transferred into a new glass bottle and mixed with Na2SO4, followed by centrifugation (at 25 °C for 3 min at 800 g). The supernatant contained the palmitoylated EWI2 cytoplasmic domain peptide and was air-dried at room temperature overnight. For overlay experiments, the peptide was first dissolved in dimethylformamide as the stock solution and further diluted with 3 % BSA-containing TBS/Tween 20 solution to working concentrations. The palmitoylation of the EWI2 cytoplasmic domain peptide was confirmed by electronspray ionization MS.
Liposome pull-down assay
Liposomes (small unilamellar vesicles) were prepared as described previously [19,20]. Briefly, POPC (1-palmitoyl-2-oleoyl phosphatidylcholine) (Avanti Polar Lipids), L-α-PtdIns4P (Avanti Polar Lipids) and 1-palmitoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphatidylcholine (16:0–12:0 NBD PC) (Avanti Polar Lipids) were added into a glass tube in 95 % chloroform/5 % methanol at a molar ratio of 53:1:6 or a mass ratio of 40:1:5 (8:0.2:1 mg). The lipids were dried under nitrogen, rehydrated in 3 ml of vesicle buffer [10 mM Tris/HCl (pH 8.0) and 150 mM NaCl] and then sonicated (at 50 Hz for 7~15 min). The sonicated vesicles were spun down in a microcentrifuge at 15000 g at room temperature for 2 h, and the liposomes were present in the upper layer of clear solution and were collected in a new tube.
To prepare EWI2 peptide-immobilized beads, 25 µl of 50 % avidin-conjugated agarose beads (Vector Laboratories) were incubated with 2 µl of 10 µg/µl biotinylated EWI2 cytoplasmic domain peptide in vesicle-binding buffer at 4°C overnight; followed by three washes with vesicle-binding buffer to remove unbound peptides. The peptide-immobilized beads were incubated with the PtdIns4P-containing fluorescent liposomes at room temperature for 1 h and then collected with brief centrifugation at low speed, followed by three washes with vesicle binding buffer. After the bound lipids were solubilized from the beads with the vesicle-binding buffer containing 0.2 % Triton X-100, the fluorescence intensity of the liposomes was measured by HTS7000 Fluoremeter (PerkinElmers) at an excitation wavelength of 485 nm and an emission wavelength of 535 nm.
Analysis of protein palmitoylation
Formetabolic labelling with [3H]palmitate, NIH 3T3 transfectants were incubated with 300 µCi/ml [9,10-3H]palmitic acid (PerkinElmer) in DMEM containing 2 % FBS at 37°C for 3 h. The cells were lysed in a 1 % Nonidet P40 lysis buffer containing 1 % Nonidet P40 (Sigma), 25 mM Hepes, 150 mM NaCl, 5 mM MgCl2, 1 mM PMSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 2 mM sodium orthovanadate and 2 mM sodium fluoride. Insoluble material was removed by centrifugation at 14000 g for 15 min and the lysates were precleared by incubation at 4°C for 6 h with Protein A– and G–Sepharose beads (Amersham Pharmacia). Then the mAb-preabsorbed Protein A– and G–Sepharose beads were incubated with cell lysate overnight at 4°C. Immune complexes were collected by centrifugation (at 4°C for 5 s at 1000 g) followed by four washes in the immunoprecipitation buffer. Immune complexes were eluted from the beads with Laemmli sample buffer and then analysed by SDS/PAGE (11%) under non-reducing conditions. The gels were treated with autoradiography enhancement reagent (PerkinElmer) according to the manufacturer’s protocol. The gels were dried and exposed to BioMax MR film (Kodak) at −80 °C for 14 days.
The palmitoylation stoichiometry of EWI2 proteins was evaluated through EWI2 dimer formation. For the effect of 2BP (2-bromopalmitic acid) on EWI2 palmitoylation, cells in culture were treated with or without 2BP (Sigma) at 100 µM overnight, then washed with PBS and lysed with 1 % Triton X-100 lysis buffer on ice for 1 h. The lysate was centrifuged at 13 000 g at 4 °C for 10 min to remove the insoluble material and then analysed by Western blot for EWI2 proteins using the anti-EWI2 mAb 5E8. For the effect of hydroxylamine (NH2OH) on EWI2 palmitoylation, the cells were lysed as described above, and the lysate was divided into two aliquots: one aliquot was treated with freshly prepared NH2OH (Pierce) (pH 7.4) at a final concentration of 1 M at 4°C overnight, and the other aliquot was treated with an equal volume of PBS as a control, followed by Western blot analysis for EWI2 proteins using the anti-EWI2mAb 5E8.
Wide-field and confocal fluorescent microscopy
The cells that were either spread on extracellular matrix-coated glass coverslips in serum-free DMEM or grown on coverslips in 10% FBS-containing DMEM were fixed with 3% paraformaldehyde at room temperature for 10 min, permeabilized with 0.1% Brij 98 at room temperature for 2–5 min, blocked with 20% goat serum at room temperature for 1 h or at 4° C overnight, and then incubated with primary mAbs at room temperature for 30 min, followed by staining with a rhodamine-conjugated goat anti-mouse IgG secondary antibody at room temperature for 30 min. After each of the antibody incubations, the cells were washed with PBS five times; each wash lasted 15 min. For double staining, the cells were further stained with Alexa Fluor® 488- or Alexa Fluor® 594-conjugated human anti-EWI2 mAb 8A12. For F-actin (filamentous actin) staining, the cells were simply incubated in Texas Red-phalloidin at room temperature for 30 min after the pretreatments, followed by extensive washes. After staining, coverslips were mounted on to glass slides in FluroSave reagent (Calbiochem).
Immunostaining of cellular PtdIns4P was based on a protocol described previously [21]. Although the protocol was reported to be able to detect the plasma membrane PtdIns4P, the present study found that intracellular PtdIns4P can also be easily detected using this protocol. Briefly, the cells were cultured on glass coverslips overnight, fixed with 3% paraformaldehyde and 0.2% glutaraldehyde at room temperature for 15 min, rinsed three times with PBS containing 50 mM NH4Cl and then chilled on ice. All subsequent steps were performed on ice with prechilled solutions. The cells were blocked and permeabilized for 45 min with a solution that contained PBS or Pipes buffer (20 mM Pipes, 137 mM NaCl and 2.7 mM KCl, pH 6.8), 5% goat serum, 50 mM NH4Cl and 0.5% saponin. The cells were incubated sequentially with anti-EWI2 mAb (IgG) overnight, Alexa Flour® 594-conjugated anti-mouse IgG secondary antibody for 1 h, PI4P mAb (IgM) for 1 h and FITC-conjugated anti-mouse IgM secondary antibody for 1 h. Each antibody incubation was followed by three washes with the Pipes buffer.
For wide-field fluorescent microscopic analysis, the cells were examined with a Zeiss Axiophot fluorescent microscope (Carl Zeiss), and the images were captured using an Optronics digital camera (Southern Micro Instrument) at a magnification of 63×. For the confocal fluorescent microscopic analysis, the coverslips were examined as described previously [22] using either a Zeiss LSM 510 laser-scanning microscope (Carl Zeiss) equipped with a 100×/1.4 Plan-APOCHROMAT oil-immersion objective or Bio-Rad 1024 confocal microscope (Bio-Rad Laboratories) equipped with a 60× oil-immersion objective. GFP, FITC or Alexa Fluor® 488, and Alexa Fluor® 594, rhodamine or Texas Red were excited by the 488 and 543 nm lines of lasers respectively, and individual channels were scanned in a series of X–Y planes to prevent cross-channel bleed-through. Each image represented a ~0.5 µm Z optical section of the cells. To collect images, similar parameters were set for different cell groups in each experiment. Images were quantified using Zeiss LSM 510 software with the thresholds set at 170 for both green and red channels. In each experiment, more than 70 cells were analysed for each transfectant, and the experiments were performed at least three times independently.
Immunoprecipitation and Western blotting
Immunoprecipitations were carried out as described previously [23]. Briefly, NIH 3T3 transfectant cells were lysed with lysis buffer at 4°C for 1 h. The lysis buffer contained 1% Brij 97 or 1% Nonidet P40 (Sigma), 150 mM NaCl, 25 mM Hepes, 2 mM PMSF 10 µg/ml leupeptin, 10 µg/ml aprotinin, 2 mM sodium vanadate and 2 mM sodium fluoride. In some experiments, the cell surface was labelled with 0.5 mg/ml EZlink sulfo-NHS (N-hydroxysuccinimido)-long-chain biotin (Pierce) in PBS at 4°C for 2 h before cell lysis. The lysates were precleared with a mixture of Protein A– and Protein G–Sepharose beads (Amersham Pharmacia) three times at 4°C after removing the insoluble material by centrifugation at 14000 g for 15 min at 4°C. The primary mAb preabsorbed Protein A– and G– Sepharose beads were then incubated with cell lysate for 3 h at 4°C. The precipitates were washed with the lysis buffer three times, dissolved in Laemmli sample buffer, heated at 95°C for 5 min, separated by SDS/PAGE (11 °C) gel and then electrically transferred on to nitrocellulose membranes (Schleicher & Schuell). For the biotinylation experiments, the membranes were blotted with HRP-conjugated extravidin (Sigma), followed by chemiluminescence (New England Nuclear Life Science) according to the manufacturer’s instructions. For Western blotting, the cell lysates were separated by SDS/PAGE (12%) gel followed by electric transfer on to nitrocellulose membranes. The nitrocellulose membranes were blocked with 5% dried milk in 1% (v/v) PBS/Tween 20 buffer, sequentially blotted with a primary mAb and HRP-conjugated anti-mouse IgG (Sigma) and detected with chemiluminescence.
Cell migration assay
Cell movement was assessed by a wound-healing assay as described previously [24]. Briefly, NIH transfectant cells were cultured in Lab-Tek Chamber slides (Nunc). After the monolayers were formed, wounds were generated by scraping the monolayers with sterile pipette tips. The detached cells were rinsed away with PBS, and the wounded monolayers were replenished with complete medium. After culture at 37 °C for 16–18 h, the monolayers were fixed and photographed under an inverted light microscope (Olympus).
Cell proliferation assay
The anchorage-dependent cell proliferation assay for NIH 3T3-EWI2 transfectants was performed under the aforementioned cell culture conditions. After being detached with trypsin-EDTA and resuspended in culturing medium, the cells were seeded in 10 cm dishes at a concentration of 2 × 105 cells per well in 10 ml of complete medium. After 72 h, the cells were harvested with trypsinization, and the live cells were counted with a haemocytometer in Trypan Blue solution.
Cell–cell adhesion assay
Cell–cell adhesion was examined using the hanging-drop aggregation assay as described previously [25]. A single-cell suspension of 1 × 104 cells in 30 µl of complete medium was suspended as a hanging drop from the lid of a 24-well culture dish and allowed to aggregate overnight at 37°C in 5%CO2 with humidity. The corresponding wells were filled with PBS to prevent the drops from drying. To assay the resistance of cell–cell adhesion to mechanical stress, the cells were subjected to shear force by passing them ten times through a 200 µl pipette tip. The cells were photographed either before or after mechanical stress, and after shear stress the number of cells in each aggregate was counted and the surface area covered by the aggregates was measured using ImageJ software (http://rsbweb.nih.gov/ij/) to portray the degree of cell aggregation.
RESULTS
Specific interactions between the EWI2 cytoplasmic domain and PIPs
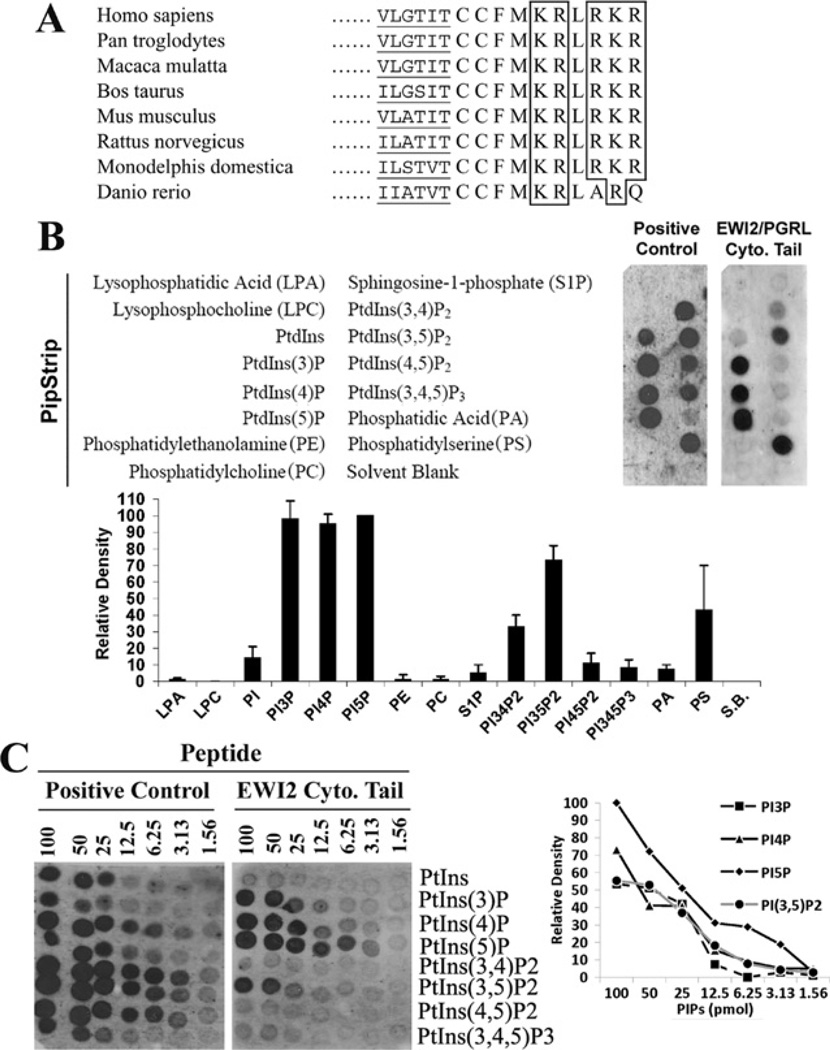
When EWI2 was identified through association with tetraspanin CD82 [3], the high number of basic residues in the short EWI2 cytoplasmic domain, five out of ten residues, was of obvious note. Importantly, these basic residues are fully conserved among mammals such as human, bovine and mouse (Figure 1A). The EWI2 orthologue of zebrafish exhibits such identity in the N-terminal basic residue cluster, but not the C-terminal one (Figure 1A). However, the amino acid sequence identity of the whole cytoplasmic tail between zebrafish and human EWI2s (80%) is still much higher than that of whole protein (38%). Because the short EWI2 cytoplasmic domain does not stretch far from the inner face of the plasma membrane that contains various acidic lipids, whether or not the positively charged EWI2 cytoplasmic tail interacts with the negatively charged lipid in the inner leaflet becomes an obvious and interesting issue. Using the protein-lipid overlay assay, we probed the lipid-immobilized membrane (PIP and Sphingo strips) with biotinylated peptides with the EWI2 cytoplasmic domain sequence. As shown in Figure 1(B), the EWI2 tail peptide only bound to some phosphoinositides, but not other membrane lipids such as phosphatidylinositol, phosphatidylcholine, phosphatidic acid or phosphatidylethanamine. The binding to phosphatidylserine was positive in some batches of PIP strips (e.g. Figure 1B), but negative in other batches (e.g. Figure 2A). For sphingolipids, the EWI2 tail bound weakly to phytosphingosine, sphingomyelin and lysophosphotidylcholine, and the positive control also bound weakly to GM1 (Supplementary Figure S1 at http://www.BiochemJ.org/bj/437/bj4370399add.htm). Among phospholipids, the EWI2 tail-binding phosphoinositides include PtdIns5P, PtdIns4P, PtdIns3P and PtdIns(3,5)P2 (phosphatidylinositol-3, 5 biphosphate). The interactions to these phosphoinositides were reproducible in all batches of PIP strips tested in the present study. EWI2 also weakly bound to PtdIns(3,4)P2 in most of the batches of PIP strips. The EWI2 tail exhibited no or merely marginal interaction with PtdIns(4,5)P2 and PtdIns(3,4,5)P3. The order of the binding affinity of phosphoinositides to EWI2 tail was PtdIns5P > PtdIns4P and PtdIns3P > PtdIns(3,5)P2 > other phosphoinositides (Figure 1C). For example, the binding of EWI2 to PtdIns5P is approximately 8-fold higher than that to PtdIns(3,5)P.
Figure 1. EWI2 cytoplasmic domain binds specifically to PIPs.
(A) The alignment of the cytoplasmic domains of vertebrate EWI2 proteins. The amino acid sequences of the full cytoplasmic domain and a partial transmembrane domain of vertebrate EWI2s were aligned. The conserved basic amino acid residues are boxed and the partial transmembrane regions are underlined. (B) The binding of EWI2 cytoplasmic domain to PIPs. The peptide with the EWI2 cytoplasmic domain sequence and the positive control were overlaid on the PIP strip by following the manufacturer’s protocol, as described in the Materials and methods section. Because the peptide was labelled with biotin at the N-terminus, the PIP strip was blotted with HRP-conjugated avidin and the bindings were visualized with chemiluminescence. The names of lipids on the left indicate the corresponding position of these lipids in the PIP strip. All lipids immobilized on the strip were at the same molar concentrations. The positive control for the PIP strip was the GST-LL5-α PH domain. For the strip blotted with the EWI2 tail peptide, the density of each dot was quantified, subtracted by the density of the Solvent Blank (S.B.) dot, and presented as a value relative to the density of PtdIns5P, which was set arbitrarily as 100, because PtdIns5P has the highest binding affinity to the EWI2 tail peptide. (C) The binding affinity of the EWI2 cytoplasmic domain to phosphoinositides. The binding of the EWI2 cytoplasmic domain peptide and the positive control GST-LL5-α PH domain to the PIP arrays were performed as described in (B). Each strip was immobilized with 100, 50, 25, 12.5, 6.25, 3.13 and 1.58 pmol of the indicated phosphoinositides. The densities of dots were quantified as described in (B).
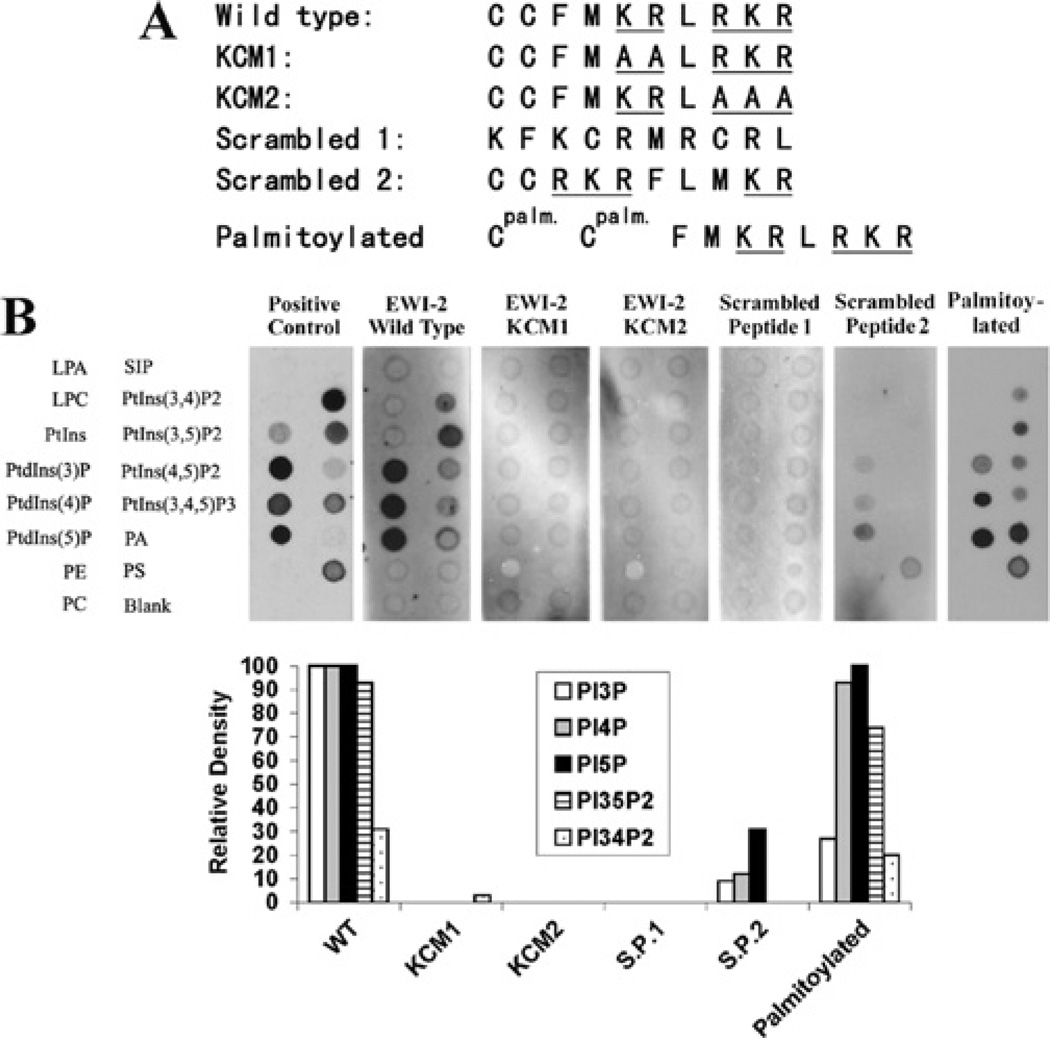
Figure 2. The basic residues in the EWI2 cytoplasmic domain determine PIP interaction.
(A) Peptide sequences of the EWI2 wild-type and mutated cytoplasmic domains are listed. In KCM1 and KCM2 mutated peptides, the two clusters of basic residues were replaced either individually or simultaneously by alanine residues, and the mutant peptides were designated as labelled. (B) All peptides were labelled with biotin at the N-termini, and the palmitoylated peptide contained a palmitate group on each cysteine residues (see the Materials and methods section). After being incubated with the peptides, the PIP strips were blotted with HRP-conjugated avidin and the bindings were visualized with chemiluminescence. The positive control is the GST-LL5-α PH domain. The densities of dots were quantified as described in Figure 1(B). LPA, lysophosphatidic acid; LPC, lysophosphocholine; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; S1P, sphingosine 1-phosphate.
To characterize the critical residues in the EWI2 cytoplasmic tail responsible for the PIP interaction, we synthesized two mutated EWI2 cytoplasmic tail peptides KCM1 and KCM2 in which the basic residue clusters were individually mutated (Figure 2A). Both KCM1 and KCM2 peptides failed to interact with PIPs (Figure 2B), indicating that the EWI2 cytoplasmic tail–PIP interaction required both basic residue clusters. We further assessed whether or not the sequence of basic residues in the EWI2 cytoplasmic tail is important for the PIP interaction. As shown in Figure 2(B), the scrambled peptide 1, which contains the amino acid composition of the EWI2 cytoplasmic tail, lost the ability to bind any PIPs. The scrambled peptide 2, in which the positions of the two basic residue clusters are inverted and become distant from each other, exhibited significantly reduced binding affinity to PIPs. To determine the effect of palmitoylation on PIP binding, the palmitoylated EWI2 tail peptide was tested in a PIP overlay assay and it was found that palmitoylation did not markedly alter the binding abilities of the EWI2 cytoplasmic tail to the aforementioned PIPs in vitro. Interestingly, the palmitoylated tail peptide gained strong binding to phosphatidic acid and bound weakly to PtdIns(4,5)P2 and PtdIns(3,4,5)P3.
Because the interaction appeared to be determined by the negative charges in phospholipids, we predicted that the polar moieties in phosphoinositides are responsible for the interaction. Surprisingly, although the EWI2 cytoplasmic tail interacted with PtdIns4P, Ins4P could not block the interactions between the EWI2 cytoplasmic tail and PtdIns4P in the protein-lipid overlay assay, even when the concentration of Ins4P was more than 100-fold compared with that of EWI2 tail peptide (Supplementary Figure S2 at http://www.BiochemJ.org//bj/437/bj4370399add.htm). As predicted, inositol-4 phosphate had no effect on the interactions of EWI2 with other PIPs. This result suggests that other structural elements in phosphoinositides are also involved in the interaction.
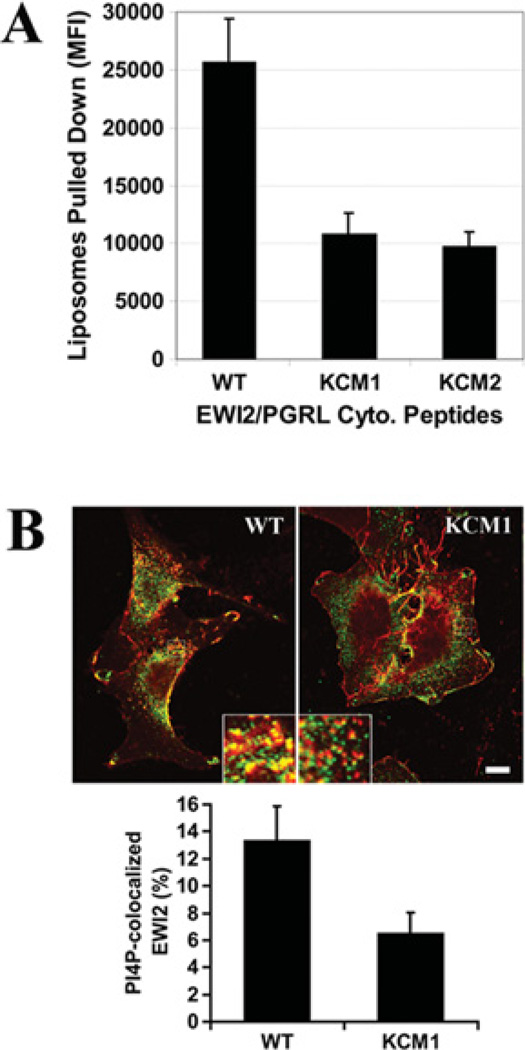
To analyse the EWI2 cytoplasmic tail and phosphoinositide interactions in a model membrane, a liposome sedimentation assay was performed. Using the fluorescent phosphatidylcholine liposomes that contain a 1.7% molar fraction of PtdIns4P, it was confirmed that the beads, immobilized with the biotinylated wild-type EWI2 cytoplasmic tail peptide, pulled-down markedly more PtdIns4P liposomes than did the KCM1 and KCM2 mutant cytoplasmic tail peptides (Figure 3A). To confirm that the PIP interaction occurred intracellularly, the co-localization of PI4P with EWI2 wild-type and KCM1 mutant proteins in NIH 3T3 transfectant cells was analysed (see below for the establishment of EWI2 transfectants; KCM2 was not analysed because its expression level was rather low). As shown in Figure 3(B), the PtdIns4P co-localization with EWI2 protein in the wild-type cells was markedly greater than that in KCM1 cells in the perinuclear area, presumably the Golgi complex region. This result is consistent with the observation from liposome pull-down experiments.
Figure 3. Interaction of the EWI2 cytoplasmic domain with membrane PIPs.
(A) The binding of the EWI2 tail to PI4P in liposomes. The fluorescent liposomes that contain POPC, PtdIns4P and NBD PC at amolar ratio of 53:1:6were incubated with the avidin-conjugated agarose beads that were immobilized with biotinylated EWI2 cytoplasmic domain peptides in vesicle-binding buffer at room temperature for 1 h. The beads were then collected with brief centrifugation, followed by three washes with vesicle-binding buffer. The fluorescence intensity of the liposomes that were pulled-down by the beads was measured at an excitation wavelength of 485 nm and an emission wavelength of 535 nm using a HTS7000 Fluoremeter (PerkinElmer). The results (means ± S.E.M.) for four independent experiments were plotted as a histogram; the differences between the wild-type and KCM1 and KCM2 peptides are statistically significant (P < 0.01). (B) The intracellular co-localization of EWI2 with PtdIns4P. NIH 3T3 transfectant cells were fixed, permeabilized and incubated with anti-EWI2 mAb, anti-PIP4 mAb and secondary antibodies as described in the Materials and methods section. Confocal microscopic images in the same Z plane were captured under the identical instrument setting. Scale bar, 10 µM. The co-localization of EWI2 (red) with PtdIns4P (green) was quantified using Zeiss LSM 510 software with a threshold of 170 set for both green and red channels. The PtdIns4P -co-localized EWI2 was presented as Manders co-localization coefficient X 100% and projected as the means ± S.E. (n = 3). In each experiment, more than 70 cells were analysed from each transfectant. P < 0.05 between wild-type and KCM1 transfectant cells.
EWI2 cytoplasmic basic residues regulate the stability of the EWI2 ectodomain
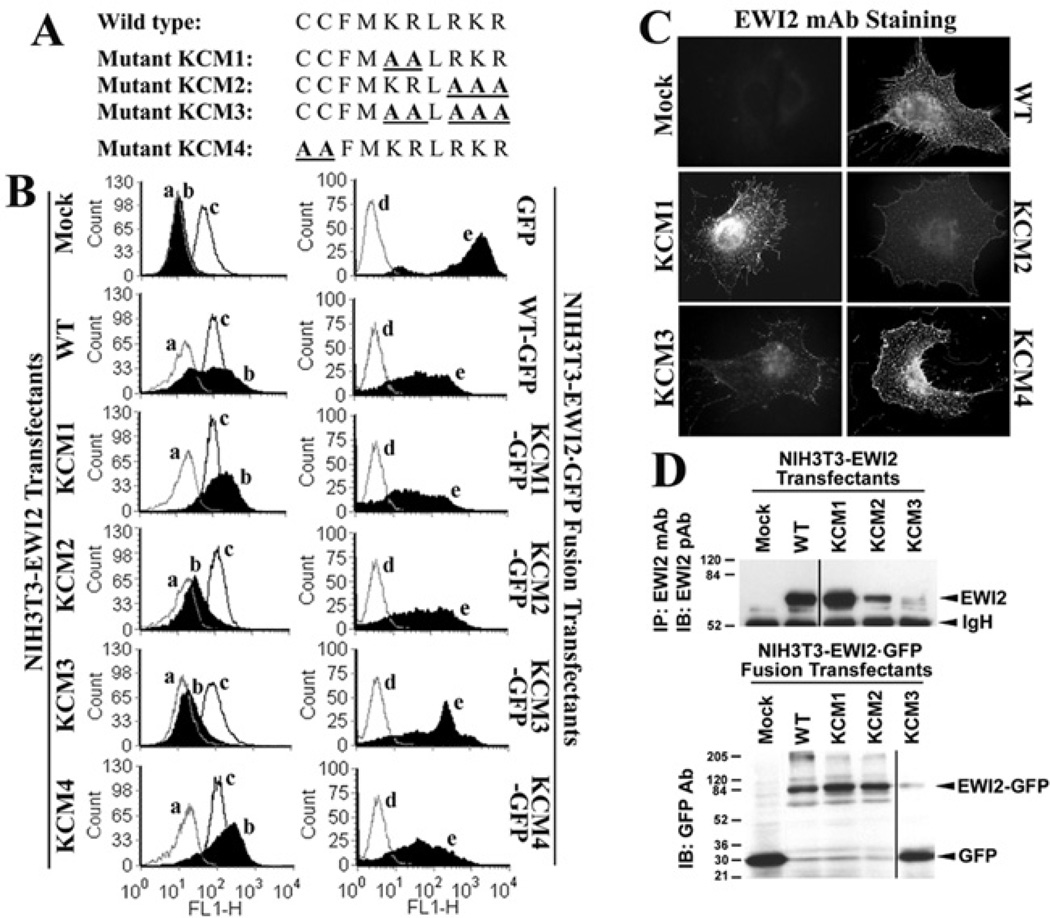
To determine the functional relevance of EWI2–phosphoinositide binding, the KR and RKR basic residue clusters in the EWI2 cytoplasmic tail were replaced with alanine residues either alone or together, and the resulting KR→AA, RKR→AAA and KRLRKR→AALAAA mutants were designated as the KCM1, KCM2 and KCM3 mutants respectively (Figure 4A). We transfected human EWI2 wild-type and KCM1, KCM2 and KCM3 mutant constructs into NIH 3T3mouse fibroblast cells and established stable transfectants by pooling and sorting hundreds of EWI2-expressing clones with flow cytometry. As shown in Figure 4(B), the KCM1 mutant could be expressed at a level equivalent to, and sometimes a little bit higher than, the wild-type on the surface of NIH 3T3 cells after flow cytometric cell sorting. The KCM2 mutant was expressed at a considerably lower level on the cell surface than were wild-type and KCM1. The expression of both mutants, especially KCM2, had the propensity to become diminished at the cell surface. The KCM3 mutant failed to express on the cell surface (Figure 4B). The expression levels of other cell-surface proteins, such as integrin α5 (Figure 4B), were equivalent on all transfectants and not significantly altered upon EWI2 expression. These results suggest that the EWI2 tail–PIP interaction is probably needed for the proper expression of EWI2 on the cell surface. To determine whether KCM2 and KCM3 mutants accumulated intracellularly and failed to reach the cell surface, we analysed the subcellular distributions of EWI2 proteins in transfectants. We found that KCM2 was not more accumulated intracellularly and the subcellular distributions of the wild-type, and KCM1 and KCM2 mutants were similar despite much less intensity of KCM2 staining (Figure 4C), suggesting that the mutations did not alter the cell-surface targeting of EWI2 proteins rather than the total expression level or stability of the proteins. The immunofluorescent signal for KCM3 was very weak, and no intracellular accumulation was found (Figure 4C), consistent with our prediction. To substantiate the finding from immunofluorescence, the total cellular level of EWI2 proteins was analysed using immunoblotting. In accordance with the results of flow cytometry and immunofluorescence, it was found that there was much less KCM2, compared with the wild-type and KCM1, and no KCM3 proteins in cell lysates (Figure 4D), confirming that both KCM2 and KCM3 mutants were not expressed inside cells either.
Figure 4. The establishment of EWI2 mutants.
(A) The schematic representation of EWI2 mutants. The mutated residues in the cytoplasmic domain of EWI2 are highlighted. (B) The expression of EWI2 wild-type and mutants in NIH 3T3 cells. The EWI2 stable transfectants were established as described in the Materials and methods section. In the left-hand panels, the NIH 3T3-Mock, -EWI2 wild-type, -KCM1, -KCM2, -KCM3 and -KCM4 transfectant cells were detached, incubated with a negative control mAb anti-mouse IgG2b, a positive control mAb anti-mouse integrin α5, and an anti-human EWI2 mAb 8A12 respectively and then stained with a FITC-conjugated secondary antibody. The stained cells were analysed by flow cytometry. The mean fluorescence intensities of EWI2 staining for NIH 3T3-Mock, -EWI2 wild-type, -KCM1, -KCM2, -KCM3 and -KCM4 mutants were 0, 306, 395, 92, 8 and 423 respectively. a, negative control; b, human EWI2; c, murine integrin α5. In the right-hand panels, the NIH 3T3 transfectants of GFP, EWI2 wild-type–GFP, KCM1–GFP, KCM2–GFP, KCM3–GFP and KCM4–GFP were analysed by flow cytometry for the green fluorescent levels (e), with mouse IgG staining as the negative control (d). (C) The subcellular distributions of EWI2 wild-type and mutants. The NIH 3T3-Mock, -wild-type, -KCM1, -KCM2 and -KCM3 transfectant cells were grown on glass coverslips for 1 day and then fixed, permeabilized and sequentially incubated with anti-EWI2 mAb and FITC-conjugated secondary antibody. The images were captured under a fluorescent microscope. Scale bar, 10 µM. (D) Total cellular protein levels of EWI2 and EWI2–GFP fusions. The NIH 3T3 transfectant cells were lysed in RIPA buffer. For EWI2 transfectants (upper panel), EWI2 proteins were immunoprecipitated with anti-EWI2 mAb 8A12 and then immunoblotted with anti-EWI2 pAb, because neither antibody detects EWI2 proteins well in Western blots. For EWI2–GFP fusion transfectants (bottom panel), the fusion proteins were analysed by Western blotting using an anti-GFP mAb. IB, immunoblot; IP, immunoprecipitation; WT, wild-type.
The failed detection of the KCM3 proteins or fewer detectable KCM2 proteins using anti-EWI2 antibodies may be caused by the disruption or vulnerability of EWI2 antigen epitopes resulting from the KCM3 and KCM2 mutations that alter the conformation of the EWI2 ectodomain. To bypass the EWI2 antigen epitopes, we also generated GFP fusions of EWI2 wild-type and mutants, in which GFPs were fused to the C-termini of EWI2s, and expressed them in NIH 3T3 cells (Figures 4B and 4D). Surprisingly, all of the fusions could be expressed at equivalent GFP levels after flow cytometric sorting (Figure 4B, right-hand panel). In Western blot analysis using anti-GFP antibodies, it was found that, unlike the wild-type–,KCM1–and KCM2–GFP fusions, the vast majority of KCM3–GFP fusions contained only the GFP moiety (Figure 4D, bottom panel). The loss of the KCM3 moiety was probably due to the improper folding of the KCM3 moiety, which led to the cleavage at the joint region of the KCM3 mutant and GFP. These results suggest that the KCM3 mutation reduced the structural stability of EWI2 proteins, leading to the degradation of EWI2 KCM3 proteins, and that the positively charged residues in the EWI2 tail probably conferred structural stability on EWI2.
Because KCM3 could not be expressed and the intact KCM2 mutant proteins were either expressed at a significantly lower level or structurally unstable, we did not include KCM2 and KCM3 in the following experiments, in which we used only the KCM1 mutant as the example for the disruption of the EWI2–PIP interaction.
EWI2 cytoplasmic basic residues are not needed for the associations between EWI2 and tetraspanins
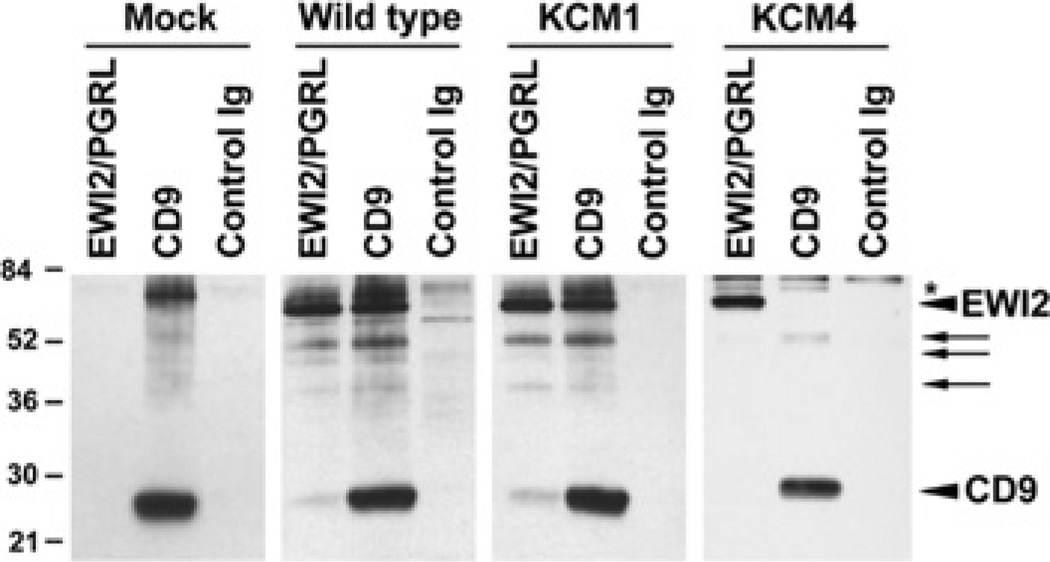
Since EWI2 associates with tetraspanins such as CD9, CD81 and CD82 [1–4], disrupted EWI2–membrane phospholipid interaction may result in aberrant association with tetraspanins. Thus the effect of the loss of the EWI2–membrane phospholipid interaction on the EWI2–tetraspanin association was assessed. Because CD9 is a major tetraspanin in NIH 3T3 cells, the EWI2–CD9 association between EWI2 wild-type and KCM1 transfectants was compared. From EWI2 immunoprecipitation profiles of surface-biotinylated NIH 3T3 cells (Figure 5), it was found that EWI2 co-precipitated with CD9 equally well in EWI2 wild-type and KCM1 transfectants. Co-precipitated CD9 was confirmed by immunoblotting (results not shown), indicating that the mutation of these basic residues in the EWI2 cytoplasmic tail did not alter the association of EWI2 with cell-surface tetraspanins and that the EWI2–phospholipid interaction was not required for EWI2–tetraspanin complex formation. Because of the unavailability of murine anti-CD81 and anti-CD82 mAbs for immunoprecipitation, we did not determine the EWI2–CD81 and –CD82 associations.
Figure 5. Effects of the PIP binding and palmitoylation of EWI2 on EWI2–tetraspanin associations.
The 1% Brij 97 lysates of the cell-surface biotinylated NIH 3T3 transfectants were immunoprecipitated with human anti-EWI2 mAb, murine anti-CD9 mAb and a control mAb. The immunoprecipitates were resolved by non-reducing SDS/PAGE and blotted with extravidin. The arrowheads indicate human EWI2 or murine CD9 proteins, arrows indicate the possibly degraded EWI2 proteins and an asterisk indicates the position of the probable endogenous murine EWI2 in CD9 precipitates.
Palmitoylation of EWI2
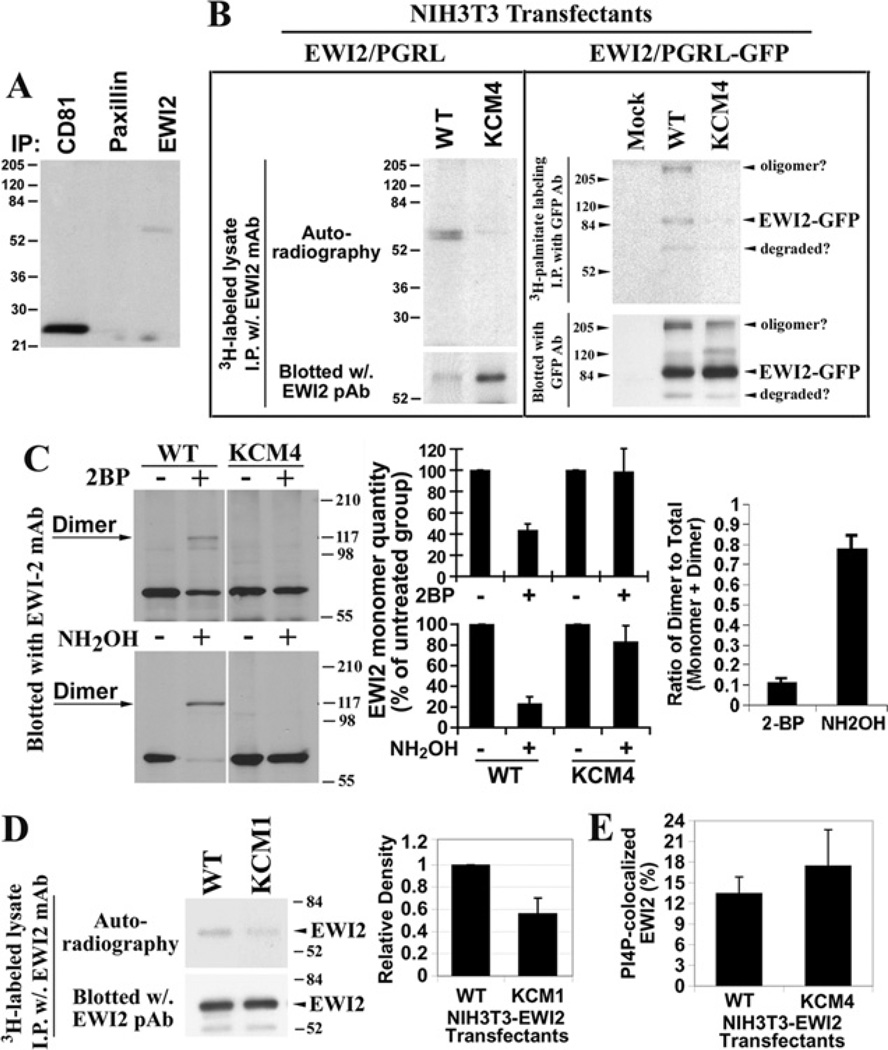
For transmembrane proteins, palmitoylation typically occurs at the cysteine residues situated adjacent to the interface of the inner leaflet of the plasma membrane lipid bilayers [26,27]. Because EWI2 contains two cysteine residues (Cys604 and Cys605) in its cytoplasmic domain near the transmembrane domain (Figure 1A), it was predicted that EWI2 could be palmitoylated, like its associated tetraspanins CD9, CD81 and CD82. EWI2 proteins were immunoprecipitated from [3H]palmitate-labelled Du145 human metastatic prostate cancer cells. It was found that EWI2 was indeed labelled by [3H]palmitate, indicating that EWI2 was constitutively palmitoylated (Figure 6A). As earlier reported, tetraspanin CD81 was heavily labelled with palmitate, and it was used here as a positive control. Paxillin was not palmitoylated as expected and served as a negative control in this experiment.
Figure 6. The palmitoylation of EWI2 and effect of the PIP interaction on palmitoylation.
(A) EWI2 is constitutively palmitoylated. Du145 prostate cancer cells were labelled with [3H]palmitate and lysed in 1% Nonidet P40 lysis buffer. EWI2, CD81 and paxillin proteins were immunoprecipitated using their specific mAbs. The precipitated proteins were resolved using non-reducing SDS/PAGE, and the labelled proteins were visualized by autoradiography. (B) Mapping the palmitoylation sites of EWI2. NIH 3T3-EWI2 wild-type and KCM4 transfectant cells were metabolically labelled with [3H]palmitate. Cells were lysed and immunoprecipitations were performed with an anti-EWI2 mAb. Half of the immunoprecipitate was resolved by SDS/PAGE and the extent of [3H]palmitate labelling was determined by autoradiography. Another half of the immunoprecipitate was analysed by EWI2 immunoblotting after SDS/PAGE separation and served as a loading control to demonstrate that equal amounts of EWI2 were immunoprecipitated under the indicated experimental conditions. (C) The induction of EWI2 covalent dimerization. The cells were treated with 2BP (100 µM) or NH2OH (1 M), and then the cell lysates were analysed by Western blot using an anti-EWI2 mAb. The levels of monomers from three independent experiments were quantified via densitometry and shown in histograms as the percentage of the ones from the untreated group of the same transfectant (middle panel). The ratios of dimer to total proteins (monomer and dimer) upon 2BP and NH2OH treatments were quantified as described above (right-hand panel). (D) The effect of the EWI2 tail–PIP interaction on EWI2 palmitoylation. In the left-hand panel, EWI2 proteins in the wild-type and KCM1 transfectants were immunoprecipitated from the [3H]palmitate-labelled cell lysate and analysed as described above. Molecular masses are given in kDa. On the right-hand pane the palmitoylated EWI2 wild-type and KCM1 proteins were quantified by measuring the band density using densitometry analysis and presented as the relative densities compared with the density of the wild-type. The results from four independent experiments were presented as the means ± S.E.M. in the histogram. P < 0.05 between wild-type and KCM1 mutant. (E) The effect of EWI2 palmitoylation on intracellular EWI2 and PtdIns4P co-localization. The experiments were performed, and the results were quantified as described in the legend for Figure 3(B). P > 0.05 between wild-type and KCM4 mutant. WT, wild-type.
To determine the palmitoylation site(s), the Cys604 and Cys605 in the EWI2 cytoplasmic tail were replaced with alanine residues, and the resulting CC→ AA mutant was designated as the KCM4 mutant (Figure 4A). We transfected human EWI2 wild-type and KCM4 mutant DNA into NIH 3T3 cells and established stable transfectants as described above. As shown in Figure 4(B), the EWI2 KCM4 mutant was stably expressed on the cell surface at levels comparable with those of the wild-type and the KCM1 mutant. After immunoprecipitating EWI2, it was found that palmitoylation was grossly diminished in the KCM4 mutant compared with that in wild-type (Figure 6B, left-hand panel). This observation was confirmed by using the GFP fusion protein of the KCM4 mutant (Figure 6B, right-hand panel). Take together, these results indicated that Cys604 and Cys605 in the EWI2 cytoplasmic tail were the palmitoylation sites. These results also strongly suggest that, unlike the membrane PIP binding, palmitoylation is not required for the cell-surface expression of EWI2 proteins.
To assess the stoichiometry of palmitoylation for EWI2 proteins, we tried the ABE (acylbiotin-exchange) method [28,29]. However, this assay did not work for EWI2 proteins. Subsequently, covalent dimerization was used to estimate the stoichiometry of EWI2 palmitoylation. Hydroxylamine specifically cleaves the thioester bond [30] and removes the palmitate groups from the intracellular cysteine residue(s) of EWI2. Because two intracellular cysteine residues in an EWI2 molecule are next to each other, the hydroxylamine-reduced cysteine residue(s) from one EWI2 molecule must form a disulfide bond(s) with only the one(s) from another EWI2 molecule immediately adjacent upon, presumably, the oxidization of lysates after cell lysis, leading to covalent dimer formation (Figure 6C). In control treatments (i.e. treated with PBS), no covalent dimer was formed (Figure 6C), underlining that EWI2 molecules are substantially palmitoylated inside cells and/or unpalmitoylated EWI2 molecules, if any, are not close enough to form a disulfide bond. Hence the dimer formation is proportional to palmitate removal. By measuring the level of dimer formation or monomer loss, we estimated the stoichiometry of palmitoylation. The formation of EWI2 dimers through the intracellular cysteine residues is confirmed by the fact that no dimer was found in the KCM4 mutant (Figure 6C). EWI2 palmitoylation appears to be highly efficient because a high proportion of EWI2/PGRL dimerized after hydroxylamine treatment, presumably because the cysteine residues freed by deacylation can oxidize and form disulfide bonds. In addition, 2BP treatment resulted in dimer formation (Figure 6C), further assuring that the intracellular cysteine residues responsible for dimer formation were palmitoylated. Both hydroxylamine and 2BP treatments reached maximal effects under the conditions described herein, based on the reduction of EWI2 monomer. Quantification of the ratio of dimer to total EWI2 proteins revealed that approximately 78% or 11% of EWI2 molecules become dimerized upon hydroxylamine or 2BP treatment respectively (Figure 6C, right-hand panel).
The effect of palmitoylation on the EWI2-tetraspanin association was then analysed. As shown in Figure 5 (right-hand panel), the association between EWI2 and CD9 was almost completely diminished in the EWI2 KCM4 mutant, although equivalent amounts of EWI2 or CD9 proteins were immunoprecipitated from KCM4 cells compared with those from wild-type and KCM1 cells. This observation indicates that palmitoylation is needed for the EWI2–tetraspanin interaction.
The PIP–EWI2 cytoplasmic tail interaction may allosterically regulate palmitoylation
The effect of the EWI2 cytoplasmic tail–PIP interaction on EWI2 palmitoylation was examined next. Using the EWI2 KCM1 mutant, which contains a mutation deficient in PIP binding in vitro, the palmitoylation level of EWI2 proteins in this mutant was compared with the palmitoylation level in the EWI2 wild-type transfectant, and it was found that the palmitoylation of EWI2 proteins was decreased significantly in KCM1 mutant (Figure 6D). These results indicate that the EWI2 cytoplasmic tail/PIP interaction is not required for the palmitoylation of EWI2, but regulates palmitoylation.
Reciprocally, palmitoylation is apparently not required for, or appears not to alter, the EWI2 cytoplasmic tail–PIP interaction in vitro, as mentioned above (Figure 1B). In addition, the co-localization of EWI2 with PtdIns4P in cells is equivalent for both wild-type and KCM4 transfectants (Figure 6E).
The functional significance of PIP binding and palmitoylation of EWI2
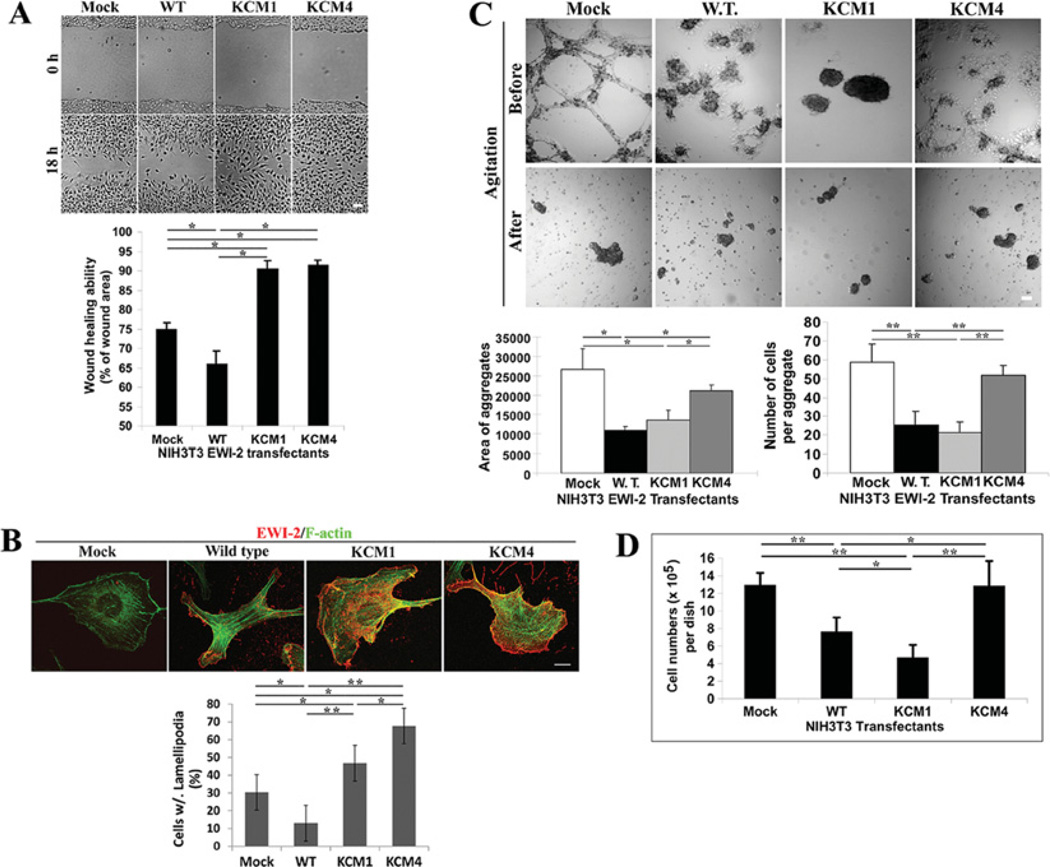
Although EWI2 associates with tetraspanins, the biological function of EWI2 remains elusive. Since EWI2 has been implicated in regulating cell spreading [16] and cell migration [3,12,13], we analysed the effects of EWI2 phospholipid binding and palmitoylation on cell migration. We measured the wound-healing abilities of NIH 3T3-EWI2 transfectants. As shown in Figure 7(A), the expression of wild-type EWI2 in NIH 3T3 cells significantly reduced collective cell migration or wound-healing ability, consistent with the negative impact of EWI2 on cell motility observed previously in other cell-movement assays [3,12,13]. Compared with EWI2 wild-type and mock transfectants, the EWI2 KCM1 and KCM4 mutants exhibited enhanced cell migration or the complete or almost complete healing of wounds after 18 h (Figure 7A). These results indicate that both cytoplasmic basic residues and palmitoylation are needed for the motility-inhibitory activity of EWI2. Because wound healing was assayed for 16–18 h and the doubling time of NIH 3T3 cells typically is longer than 20–24 h, the contribution of cell proliferation to wound healing is negligible. To substantiate this observation, we investigated the effect of EWI2 expression on the motility-related subcellular morphology and cytoskeletal organization. As shown in Figure 7(B), EWI2 wild-type largely inhibited the formation of lamellipodia, whereas lamellipodia in the mutant cells, especially in KCM4 cells, were much more frequently seen and became robust, even exceeding the ones seen in mock cells. In terms of actin cytoskeleton, more transverse stress fibres were found in wild-type cells, whereas a better-developed cortical meshwork was observed in KCM1 and KCM4 cells (Figure 7B). Such results correlate well with the wound-healing abilities of these transfectants.
Figure 7. The PIP interaction and palmitoylation of the EWI2 cytoplasmic tail regulates cell migration, cell–cell adhesion and cell proliferation.
(A) NIH 3T3-Mock, -EWI2 wild-type, and -KCM1 and -KCM4 transfectant cells were seeded in six-well culture plates at confluence in complete medium for 4 h before the wounds were generated and then further cultured for 18 h after the wounds were generated. Scale bar, 100 µM. The experiments were repeated five times and the representative results from one experiment are shown. The wounded and healed areas were quantified with the NIH/Scion Imaging analysis tool. Cell migration is expressed as the healed area as a percentage of the wounded area. The results were presented as means ± S.E.M. *P <0.05. (B) The formation of lamellipodia and stress fibres in NIH 3T3-EWI2 transfectants. The cells were cultured in complete media for 2 days before fixation, permeabilization and immunoflurescent staining as described in the Materials and methods section. The fluorescent images of the basal XY sections were collected under a confocal microscope. Lamellipodia were identified by anti-EWI2 mAb staining at the cell periphery and stress fibres were identified by α-phalloidin staining. Scale bar, 10 µM. The cells containing lamellipodia were counted and compared statistically between transfectants. **P < 0.01; *P < 0.05. (C) Cell aggregation of NIH 3T3-EWI2 transfectant cells was assayed in the hanging drops of complete media. The morphology of aggregates before and after shear stress is presented in the light microscopic images (scale bar, 100 µM), and the aggregation was quantified as the total surface area covered by aggregates and the average number of cells per aggregate. *P < 0.05. *P < 0.05. (D) Anchorage-dependent cell proliferation. As described in the Materials and methods section, NIH 3T3-Mock, -EWI2 wild-type, -KCM1 and -KCM4 transfectants were seeded at the same numbers in regular cell culture dishes, cultured for 72 h and quantified after detachment. The results represent the means ± S.E.M. of the results from four independent experiments. In each experiment, each transfectant was assayed in duplicate. **P < 0.01; *P < 0.05. WT/W.T., wild-type.
Wound healing is collective cell migration or a cell-migratory process that is directly regulated by cell–cell adhesion. For example, the retardation of wound healing could result from either suppressed cell migration or increased cell–cell adhesion that resists the scattering of closely apposed cells in monolayer. In addition, as an IgSF protein, EWI2 probably functions as a cell–cell adhesion molecule. Consistently, EWI2 is located at cell–cell contacts [4], implicating its involvement in cell–cell adhesion. In particular, identification of the cell-bound heat-shock protein A8 as a ligand of EWI2 [14] strengthens this notion. Hence, the roles of EWI2 wild-type and mutants in cell–cell adhesion was assessed by analysing cell aggregation. As shown in Figure 7(C), before shear stress, the mock and KCM4 mutant cells formed web- or interconnected cable-like aggregates in hanging drops, whereas the wild-type and KCM1 mutant cells could not form or maintain web-like aggregates. The partially or fully disrupted web-like aggregates reflect deficient cell–cell adhesion that cannot resists cellular intrinsic traction (results not shown). After shear stress which disrupts web-like aggregates, the degrees of cell aggregation were quantified and it was found that the wild-type and KCM1 mutant cells exhibited obviously reduced cell aggregation compared with mock and KCM4 cells (Figure 7C, bottom panel), consistent with their morphological phenotypes in hanging drops before shear stress.
Since cell–cell adhesion modulates cell proliferation in a contact-inhibition manner, we then determined whether EWI2 regulates cell proliferation. For anchorage-dependent cell proliferation, EWI2 expression markedly inhibited the proliferation of NIH 3T3 cells (Figure 7D). Notably, the KCM1 mutant potentiated the inhibitory activity, whereas the KCM4 mutant fully lost it (Figure 7D), strongly suggesting that the basic residues of EWI2 alleviate while the palmitoylation of EWI2 is required for the inhibition of cell growth.
DISCUSSION
The selective binding of the EWI2 cytoplasmic domain to phosphoinositides require both electrostatic and conformational interactions
The major breakthrough in understanding the mechanisms by which proteins interact with membrane PIPs is the identification of PIP-binding motifs in proteins [31,32]. PIP-binding domains include the PH (pleckstrin homology), PX (Phox homology), ENTH (epsin N-terminal homology), FYVE and C2 [PKC (protein kinase c) conserved region 2] domains [31,32]. In addition, the protein–PIP binding involves other structural determinants such as tubby motif and basic residue clusters [32,33]. The basic residue clusters typically mediate the PIP interactions of cytosolic proteins such as MARCKS (myristoylated alanine-rich C-kinase substrate) and ERM (ezrin/radixin/moesin) proteins [32,34,35]. The present study reveals that the basic residue clusters can also mediate the interaction between the cytoplasmic domain of a transmembrane protein and PIPs, which was reported only for the binding of the synaptotagmin C2 domain to PtdIns(4,5)P2 [33].
Beyond the charged interaction, the sequence of basic residues and position of basic residue clusters in the EWI2 tail are important for the PIP interaction (Figure 2A). Because the surrounding hydrophobic residues can reinforce the interaction via partial penetration into the lipid bilayer [36,37], the adjacent hydrophobic residues in the EWI2 tail such as leucine/phenylalanine and methionine may also play a role in the interaction. Reciprocally, the conformation of PIPs also appears to be required for the EWI2–PIP interaction, based on the selective interaction of the EWI2 tail peptide with PtdIns(3,5)P2, but not with PtdIns(4,5)P2 and PtdIns(3,4)P2, and the higher binding affinity to monophosphoinositides compared with biphosphoinositides. A phosphate at the 5′ position of the inositol ring seems to confer the optimal conformation to interact with the EWI2 tail. It is likely that the EWI2 tail forms a positively charged pocket that is best suited for accommodating one phosphate group. For the EWI2 and biphosphoinositide interaction, the phosphate at the 4′ position of the inositol ring is so close to the one at the 5′ position of the ring that it may impose a steric hindering effect on the interaction of the 5′ phosphate with the positively charged region in the EWI2 tail.
Phosphoinositides are differentially localized in subcellular compartments [38]. For example, the cellular distributions of PtdIns3P, PtdIns4P and PtdIns(3,5)P2 are limited mainly to the endocytic vesicle, Golgi complex and multivesicular bodies respectively [32,33,38]. Thus the interactions of EWI2 with PtdIns4P, PtdIns3P and PtdIns(3,5)P2 are more likely to occur in the maturation of EWI2 proteins and/or sorting of EWI2 to the microdomains enriched in these phospholipids. For example, the binding to PtdIns4P in the Golgi complex may help retain the structural stability of EWI2, consistent with our observation that more co-localization of EWI2 with PtdIns4P at the perinuclear region, where the Golgi complex situates, correlates with better stability of EWI2 proteins (Figure 3B). Of note, EWI2 is one of the few PtdIns5P-binding proteins, which include SNX13 and ING2 [33], reported so far. However, PtdIns5P is not a major PIP in mammalian cellular membranes, and its cellular level is quite low compared with that of other PIPs [33]. Also, its subcellular localization remains obscure and, presumably, it is present in the plasma membrane and late endosomes [32,33]. Although PtdIns(4,5)P2 is the major PIP in the inner leaflet of the plasma membrane, most PtdIns(4,5)P2 in the plasma membrane is generated from PtdIns4P, but not PIP [32,38]. Thus the physiological relevance of PtdIns5P binding remains to be determined.
Tetraspanins associate with type II PtdIns4 kinase and activated PKC in the context of TEMs (tetraspanin-enriched microdomains) [39,40]. PtdIns4P, the product of PtdIns4 kinases, is probably more concentrated in TEMs than in other parts of the plasma membrane. As a component of TEMs, EWI2 may have more chances to bind to PtdIns4P than to other phosphoinositides. Based on the recent finding that PtdIns4P actually exists abundantly at the plasma membrane [21], PtdIns4P is more likely to be the major PIP that EWI2 binds at the plasma membrane or even in an entire cell, especially given the fact that EWI2 is localized mainly at the plasma membrane [4,13]. Functionally, PtdIns4P may facilitate EWI2 to be targeted into and associate with TEMs. But the binding of PIPs appears not to be essential for the EWI2–tetraspanin association, indicating that other structural elements in EWI2 are needed for its interaction with TEMs. Another possibility is that EWI2 may sequester PtdIns4P in TEMs, which is in turn converted into PtdIns(4,5)P2 and results in a higher concentration of PtdIns(4,5)P2 in TEMs. A local enrichment of diacyglycerol, derived from PtdIns(4,5)P2 during the activation of PKC, in TEMs provides one of the structural bases for the regulated TEMs–PKC association.
Besides phosphoinositides, the EWI2 tail may also bind to cytoplasmic proteins. Sala-Valdes et al. [13] found that EWI2 binds directly to ERM proteins through its cytoplasmic tail, and this ERM interaction links TEMs to the actin cytoskeleton. Although the EWI2 tail confers the structural element(s) for the ERM interaction, the requirement of the EWI2 tail for the ERM interaction was not established [13]. Nevertheless, it is unlikely for the short and positively charged EWI2 tail to associate simultaneously with ERM proteins and phosphoinositides in a direct manner. The binding of the EWI2 tail to phospholipids and ERM proteins may be mutually regulated. For example, when the EWI2-binding PIPs such as PtdIns4P become enriched at local areas of the plasma membrane such as TEMs, these PIPs may replace ERM proteins to bind EWI2 and disconnect TEMs from the actin cytoskeleton. Effects of PIP binding on the EWI2–ERM association and vice versa remain critical issues to be assessed in future studies. In addition, whether phosphatidylserine functions as an anchorage site for the EWI2 tail at the plasma membrane also needs to be addressed.
The EWI2–phospholipid interaction probably regulates EWI2 palmitoylation
Palmitoylation regulates membrane targeting, membrane compartmentalization and trafficking of proteins [27,41]. Many palmitoylated proteins contain basic residue(s) that are located near the palmitoylated cysteine residues [42]. These basic residues may promote the interaction of palmitoylated proteins with the acidic inner leaflet of the plasma membrane [42]. Although direct evidence supporting this hypothesis seems lacking, a previous study demonstrated that the palmitoylation of IgSF protein CD4 is not altered by mutation at the surrounding basic residues in the CD4 cytoplasmic tail [43]. For EWI2, phospholipid binding is apparently not required for palmitoylation. The membrane-proximal basic residue cluster actually facilitates the palmitoylation, indicating that the EWI2 tail is prone to palmitoylation when it attaches to the inner leaflet. Thus the phenotypes of the KCM1 mutant either result partially from diminished palmitoylation (see following) or are the combinatory effect of disrupted PIP binding and diminished palmitoylation.
Reciprocally, the palmitoylation of EWI2 is unlikely to be required for the phosphoinositide interaction, at least in vitro, because the unpalmitoylated EWI2 tail peptide binds phosphoinositides. Also, palmitoylation appears not to alter the EWI2–PIP interactions because the palmitoylated and unpalmitoylated tail peptides bind PIPs equally well in vitro. Accordingly we predict that the cytoplasmic tail of unpalmitoylated EWI2 still interacts with phosphoinositides in the inner lipid layer. Therefore the lipid-binding property of EWI2 is independent of palmitoylation.
EWI2 is constitutively palmitoylated as other TEM constituents, such as tetraspanins and LN-binding integrins [9,10]. It is likely that FPRP, another EWI2 Ig subfamily member that associates with TEMs, is also palmitoylated based on the presence of membrane-proximal cysteine residues in its cytoplasmic domain. Since TEM sconsist of various palmitoylated transmembrane proteins, it is biochemically the PEM (palmitate-enriched microdomain). Although the significance of such lipid organization is unclear, enriched palmitate chains are predicted to make the inner leaflet of the membrane relatively even in thickness. In addition, palmitoylation increases the volume of hydrocarbon chains in the inner leaflet of the lipid bilayer and thereby enhances the propensity of the inner leaflet to form hexagonal II phase locally [44], which may alter the membrane fluidity and permeability. Moreover, the EWI2–PIP interaction brings phospholipids into palmitoylation-enriched TEMs and leads to the lipid reorganization of TEMs. In EWI2-containing TEMs, a higher concentration of phosphoinositides and palmitates may alter the biophysical features of TEMs such as membrane thickness and curvature, as well as the biochemical function of TEMs as a signalling platform.
Differential and overlapping functions of membrane phospholipid binding and palmitoylation of the EWI2 cytoplasmic domain
We can classify the cellular functions altered by EWI2 mutations into basic residue- and/or palmitoylation-dependent. For example, the enhanced ability of wound healing is a functional alteration dependent upon both basic residues and palmitoylation of the EWI2 cytoplasmic tail. In this case, aberrancy in either positive charge or palmitoylation leads to the phenotype, indicating an essential role of the EWI2 tail in inhibiting cell migration. Since the loss of basic residues also down-regulates palmitoylation (Figure 6C), this biological effect of the KCM1 mutant could result from less palmitoylation of this mutant.
In contrast, the decreased protein stability of EWI2 was observed in only the basic residue mutants, but not in the palmitoylation mutant, and was probably caused by the disruption of charge-mediated interactions such as PIP binding and ERM protein association. The incremental instability from KCM1 and KCM2 to KCM3 mutant proteins suggests that the contributions of two basic residue clusters are different intracellularly and that the C-terminal cluster plays a more critical role inside cells. Compared with the wild-type, the KCM1 mutant behaves opposite to the KCM4 mutant in regulating cell proliferation and has no significant impact on cell–cell adhesion, in contrast with the marked effect of the KCM4 mutant, further supporting the hypothesis that the basic residues and palmitoylation of the EWI2 tail define distinct EWI2 functions. Of note, the effects of KCM1 mutation on cell adhesion and proliferation could result from loss of PIP binding, aberrant biosynthesis and/or maturation of the EWI2 protein, disruption of the interaction between the EWI2 tail and ERM protein [13], or a combination of these possibilities. Nevertheless, the present study demonstrated that the positively charged cytoplasmic residues of the EWI2 protein are required for the structural and functional integrity of EWI2.
Wound healing is a collective cell-migration process and therefore also depends on the adhesiveness between migrating cells and their surrounding cells. Compared with the mock cells, KCM1 exhibited an increase in wound closure, but reduced cell–cell adhesion, suggesting that the increased cell migration may result from reduced cell–cell adhesion. In contrast, when compared with the wild-type, KCM1 exhibited the better wound closure, but had no effect on cell–cell adhesion, suggesting that KCM1 indeed has significantly better intrinsic motility than wild-type. Relative to mock or wild-type cells, KCM4 had much better wound closure and exhibited unchanged or increased cell–cell adhesiveness respectively. Hence, the better ability of KCM4 to heal wounds results directly from their higher intrinsic motility. Taken together, relative to the wild-type, the better collective migration of the KCM1 and KCM4 mutants results from the net increase in cell motility and is independent of their cell–cell adhesiveness.
In addition, the cell–cell adhesiveness of EWI2 transfectants is well correlated with their abilities to proliferate, suggesting that the cells with shorter doubling time and/or less contact inhibition have more and/or activated cell–cell adhesion molecules. Although contact inhibition of proliferation is well recognized, the strength of cell–cell adhesion has never been analysed in conjunction with this phenomenon. The present study for the first time reveals that the strength of cell–cell adhesion can be linked to the degree of contact-mediated inhibition of proliferation.
In conclusion, the positive charge and palmitoylation of the EWI2 cytoplasmic tail regulate the same and different functional properties of EWI2–PIP binding seems to determine the stability of EWI2 proteins, palmitoylation is needed for EWI2-mediated down-regulation of cell proliferation and cell–cell adhesion, and PIP binding and palmitoylation both appear to be required for EWI2-dependent inhibition of cell migration and lamellipodia formation
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Lisa Jennings, Dr Zheng Fan and Dr David Armbruster for constructive suggestions and critical review of the manuscript, Dr Tamás Schweighoffer for reagents, and Dr Jie Zheng and Dr Zhao Wang for technical support. We thank all colleagues who provided the antibodies used in this study.
FUNDING
This work was supported by the National Institutes of Health [grant number CA-096991] and the Department of Defense Research [grant number W81XWH-04-1-0156] (to X.A.Z). E.R. is supported by Association pour la Recherche sur le Cancer.
Abbreviations used
- 2BP
2-bromopalmitic acid
- DMEM
Dulbecco’s modified Eagle’s medium
- ECM
extracellular matrix
- ERM
ezrin/radixin/moesin
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- GFP
green fluorescent protein
- HRP
horseradish peroxidase
- IgSF
immunoglobulin superfamily
- LN
laminin
- mAb
monoclonal antibody
- NHS
N-hydroxysuccinimido
- pAb
polyclonal antibody
- PH
pleckstrin homology
- PIP
phosphatidylinositol phosphate
- PKC
protein kinase C
- C2
PKC conserved region 2
- POPC
1-palmitoyl-2-oleoyl phosphatidylcholine
- TEM
tetraspanin-enriched microdomain
Footnotes
AUTHOR CONTRIBUTION
Bo He, Yanhui Zhang, Mekel Richardson and Julian Zhang conducted experiments. Xin Zhang designed the project. Eric Rubinstein provided special reagents and advice. Xin Zhang, Bo He and Yanhui Zhang wrote the paper and prepared the Figures.
REFERENCES
- 1.Clark KL, Zeng Z, Langford AL, Bowen SM, Todd SC. PGRL is a major CD81-associated protein on lymphocytes and distinguishes a new family of cell surface proteins. J. Immunol. 2001;167:5115–5121. doi: 10.4049/jimmunol.167.9.5115. [DOI] [PubMed] [Google Scholar]
- 2.Stipp CS, Kolesnikova TV, Hemler ME. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 2001;276:40545–40554. doi: 10.1074/jbc.M107338200. [DOI] [PubMed] [Google Scholar]
- 3.Zhang XA, Lane WS, Charrin S, Rubinstein E, Liu L. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. 2003;63:2665–2674. [PubMed] [Google Scholar]
- 4.Charrin S, Le Naour F, Labas V, Billard M, Le Caer JP, Emile JF, Petit MA, Boucheix C, Rubinstein E. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 2003;373:409–421. doi: 10.1042/BJ20030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charrin S, Le Naour F, Oualid M, Billard M, Faure G, Hanash SM, Boucheix C, Rubinstein E. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 2001;276:14329–14337. doi: 10.1074/jbc.M011297200. [DOI] [PubMed] [Google Scholar]
- 6.Stipp CS, Orlicky D, Hemler ME. FPRP, a major, highly stoichiometric, highly specific CD81- and CD9-associated protein. J. Biol. Chem. 2001;276:4853–4862. doi: 10.1074/jbc.M009859200. [DOI] [PubMed] [Google Scholar]
- 7.Boucheix C, Rubinstein E. Tetraspanins. Cell. Mol. Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarrant JM, Robb L, van Spriel AB, Wright MD. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 2003;24:610–617. doi: 10.1016/j.it.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 10.Hemler ME. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 11.Liu MW, Zhang XA. KAI1/CD82, a tumor metastasis suppresser. Cancer Lett. 2006;240:183–194. doi: 10.1016/j.canlet.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Stipp CS, Kolesnikova TV, Hemler ME. EWI-2 regulates α3β1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 2003;163:1167–1177. doi: 10.1083/jcb.200309113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemle ME, Sanchez-Madrid F, Yanez-Mo M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ERM proteins. J. Biol. Chem. 2006;281:19665–19675. doi: 10.1074/jbc.M602116200. [DOI] [PubMed] [Google Scholar]
- 14.Kettner S, Kalthoff F, Graf P, Priller E, Kricek F, Lindley I, Schweighoffer T. EWI-2/CD316 is an inducible receptor of HSPA8 on human dendritic cells. Mol. Cell. Biol. 2007;27:7718–7726. doi: 10.1128/MCB.00180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolesnikova TV, Kazarov AR, Lemieux ME, Lafleur MA, Kesari S, Kung AL, Hemler ME. Glioblastoma inhibition by cell surface immunoglobulin protein EWI-2, in vitro and in vivo. Neoplasia. 2009;11:77–86. doi: 10.1593/neo.81180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolesnikova TV, Stipp CS, Rao RM, Lane WS, Luscinskas FW, Hemler ME. EWI-2 modulates lymphocyte integrin α4β1 functions. Blood. 2004;103:3013–3019. doi: 10.1182/blood-2003-07-2201. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XA, Kazarov AR, Yang X, Bontrager AL, Stipp CS, Hemler ME. Function of the tetraspanin CD151-α6β1 integrin complex during cellular morphogenesis. Mol. Biol. Cell. 2002;13:1–11. doi: 10.1091/mbc.01-10-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milligan G. Signal Transduction: a Practical Approach. Oxford: Oxford University Press; 1999. [Google Scholar]
- 19.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTor signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 20.Mozafari MR. Liposomes: an overview of manufacturing techniques. Cell. Mol. Biol. Lett. 2005;10:711–719. [PubMed] [Google Scholar]
- 21.Hammond GRV, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P2. Biochem. J. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Q, Xia B, Moshiach S, Xu C, Jiang Y, Chen Y, Sun Y, Lahti JM, Zhang XA. The microenvironmental determinants for kidney epithelial cyst morphogenesis. Eur. J. Cell Biol. 2008;87:251–266. doi: 10.1016/j.ejcb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He B, Liu L, Cook GA, Grgurevich S, Jennings LK, Zhang XA. Tetraspanin CD82 attenuates cellular morphogenesis through down-regulating integrin α6-mediated cell adhesion. J. Biol. Chem. 2005;280:3346–3354. doi: 10.1074/jbc.M406680200. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, He B, Liu WM, Zhou D, Cox JV, Zhang XA. Tetraspanin CD151 promotes cell migration through regulating integrin trafficking. J. Biol. Chem. 2007;282:31631–31642. doi: 10.1074/jbc.M701165200. [DOI] [PubMed] [Google Scholar]
- 25.Qin Y, Capaldo C, Gumbiner BM, Macara IG. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 2005;171:1061–1071. doi: 10.1083/jcb.200506094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 27.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 28.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. BioTechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 29.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat. Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 30.Jimenez CR, Huang L, Qiu Y, Burlingame AL. Current Protocols in Protein Science. New York: John Wiley & Sons; 1998. [Google Scholar]
- 31.Sato TK, Overduin M, Emr SD. Location, location, location: membrane targeting directed by PX domains. Science. 2001;294:1881–1885. doi: 10.1126/science.1065763. [DOI] [PubMed] [Google Scholar]
- 32.Roth MG. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 33.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 34.Niggli V. Structural properties of lipid-binding sites in cytoskeletal proteins. Trends Biochem. Sci. 2001;26:604–611. doi: 10.1016/s0968-0004(01)01927-2. [DOI] [PubMed] [Google Scholar]
- 35.Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- 36.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 37.Balla T. Inositol-lipid binding motifs: signal integrators through protein–lipid and protein–protein interactions. J. Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 38.De Matteis MA, Godi A. PI-loting membrane traffic. Nat. Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 39.Berditchevski F, Tolias KF, Wong K, Carpenter CL, Hemler ME. A novel link between integrins, TM4SF proteins (CD63, CD81) and phosphatidylinositol 4-kinase. J. Biol. Chem. 1997;272:2595–2598. doi: 10.1074/jbc.272.5.2595. [DOI] [PubMed] [Google Scholar]
- 40.Zhang XA, Bontrager AL, Hemler ME. Transmembrane 4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific β1 integrins. J. Biol. Chem. 2001;276:25005–25013. doi: 10.1074/jbc.M102156200. [DOI] [PubMed] [Google Scholar]
- 41.Bijlmakers MJ, Marsh M. The on-off story of protein palmitoylation. Trends Cell Biol. 2003;13:32–42. doi: 10.1016/s0962-8924(02)00008-9. [DOI] [PubMed] [Google Scholar]
- 42.El-Husseini AE, Bredt DS. Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 2002;3:791–802. doi: 10.1038/nrn940. [DOI] [PubMed] [Google Scholar]
- 43.Popik W, Alce TM. CD4 receptor localized to non-raft membrane microdomains supports HIV-1 entry. J. Biol. Chem. 2004;279:704–712. doi: 10.1074/jbc.M306380200. [DOI] [PubMed] [Google Scholar]
- 44.Krönke M. Biophysics of ceramide signaling: interaction with proteins and phase transition of membranes. Chem. Phys. Lipids. 1999;101:109–121. doi: 10.1016/s0009-3084(99)00059-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.