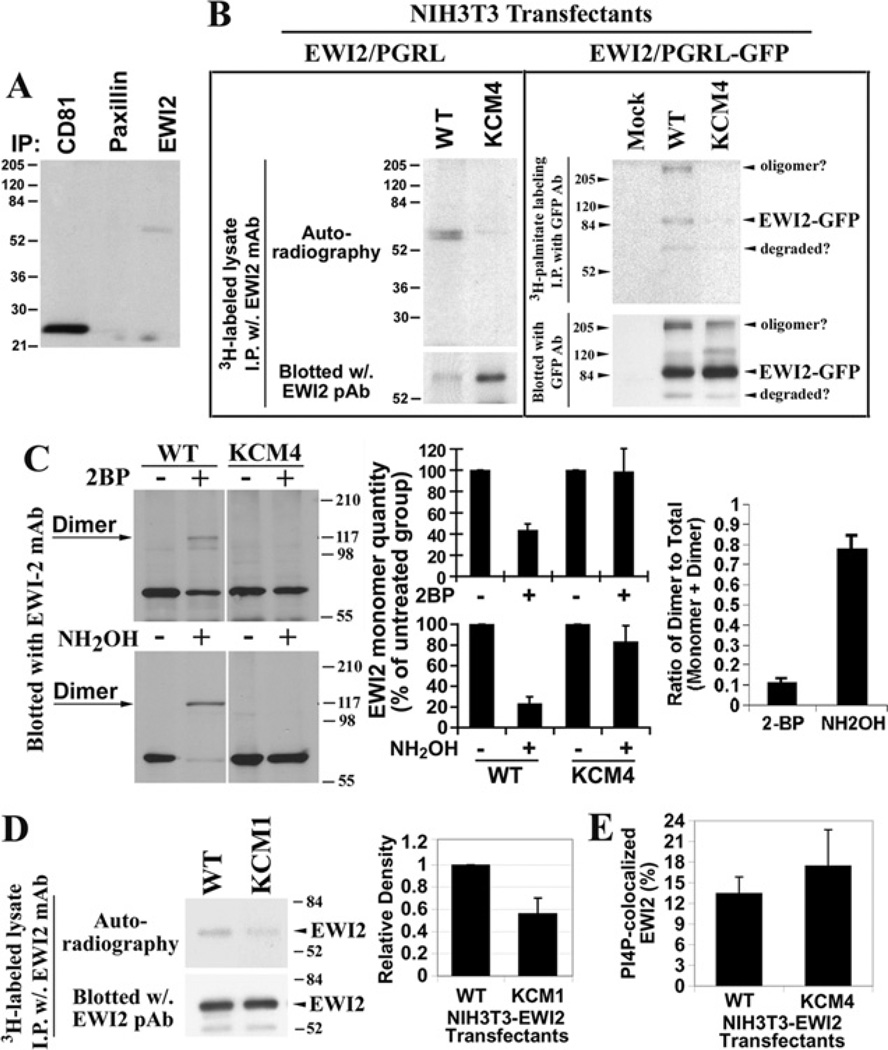
Figure 6. The palmitoylation of EWI2 and effect of the PIP interaction on palmitoylation.
(A) EWI2 is constitutively palmitoylated. Du145 prostate cancer cells were labelled with [3H]palmitate and lysed in 1% Nonidet P40 lysis buffer. EWI2, CD81 and paxillin proteins were immunoprecipitated using their specific mAbs. The precipitated proteins were resolved using non-reducing SDS/PAGE, and the labelled proteins were visualized by autoradiography. (B) Mapping the palmitoylation sites of EWI2. NIH 3T3-EWI2 wild-type and KCM4 transfectant cells were metabolically labelled with [3H]palmitate. Cells were lysed and immunoprecipitations were performed with an anti-EWI2 mAb. Half of the immunoprecipitate was resolved by SDS/PAGE and the extent of [3H]palmitate labelling was determined by autoradiography. Another half of the immunoprecipitate was analysed by EWI2 immunoblotting after SDS/PAGE separation and served as a loading control to demonstrate that equal amounts of EWI2 were immunoprecipitated under the indicated experimental conditions. (C) The induction of EWI2 covalent dimerization. The cells were treated with 2BP (100 µM) or NH2OH (1 M), and then the cell lysates were analysed by Western blot using an anti-EWI2 mAb. The levels of monomers from three independent experiments were quantified via densitometry and shown in histograms as the percentage of the ones from the untreated group of the same transfectant (middle panel). The ratios of dimer to total proteins (monomer and dimer) upon 2BP and NH2OH treatments were quantified as described above (right-hand panel). (D) The effect of the EWI2 tail–PIP interaction on EWI2 palmitoylation. In the left-hand panel, EWI2 proteins in the wild-type and KCM1 transfectants were immunoprecipitated from the [3H]palmitate-labelled cell lysate and analysed as described above. Molecular masses are given in kDa. On the right-hand pane the palmitoylated EWI2 wild-type and KCM1 proteins were quantified by measuring the band density using densitometry analysis and presented as the relative densities compared with the density of the wild-type. The results from four independent experiments were presented as the means ± S.E.M. in the histogram. P < 0.05 between wild-type and KCM1 mutant. (E) The effect of EWI2 palmitoylation on intracellular EWI2 and PtdIns4P co-localization. The experiments were performed, and the results were quantified as described in the legend for Figure 3(B). P > 0.05 between wild-type and KCM4 mutant. WT, wild-type.