Abstract
Site-specific recombination tools such as the Cre–loxP system are used to create animal models where conditional gene deletion/activation studies are required. In the current proof of principle study, we have demonstrated that a PET reporter gene (PRG), the herpes simplex virus type 1 thymidine kinase (HSV1-tk), can be made to remain silent and can be activated by Cre–loxP-mediated recombination in cell culture and in living mice. An adenovirus carrying a silent HSV1-tk was tail-vein injected (1×109 PFU) in six transgenic mice that express Cre recombinase in their liver (Cre+) and in four control mice (Cre−). The liver-specific expression of the PRG in Cre+ mice was detected in the microPET following injection of the reporter probe, 9-[4-fluoro-3-(hydroxymethyl)butyl]guanine ([18F]-FHBG). The [18F]-FHBG accumulation in the liver in terms of percent-injected dose per gram of tissue was 7.72±1.13 for the Cre+ mice and 0.10±0.02 for the Cre− mice (P<0.05) 48 h after adenoviral injection. These results were further validated by quantitative RT-PCR, western blotting and by in vitro assays for herpes simplex virus type 1 thymidine kinase enzyme activity. Thus by using the Cre–loxP system it is possible to modulate a PRG and noninvasively monitor the extent of Cre–loxP-mediated gene activation by imaging in a microPET scanner.
Keywords: Cre recombinase, Cre–loxP system, positron emission tomography reporter gene, herpes simplex virus type 1 thymidine kinase, noninvasive imaging, conditional gene activation/deletion
Introduction
The adoption of molecular biological techniques for genetic manipulation of the mouse has resulted in the generation of transgenic/knockout mouse models for the investigation of many aspects of mammalian biology. These animal models allow researchers to study the in vivo phenotypes of the specific gene that was either introduced or deleted. However, there are often instances where introduction or deletion of such a transgene into the mouse results in a survival problem in the embryo or in juvenile development. Thus, it may become difficult or impossible to maintain the transgenic mouse line by breeding. In these circumstances, it is advantageous to design a dormant gene that can be activated after the establishment of the transgenic line. This is usually achieved by the use of site-specific recombinases.1,2
Site-specific recombinases are useful reagents for genomic manipulations that allow for tissue-specific or developmental stage-specific modulation of gene expression in knockout and transgenic mice.3 The four members of the family of site-specific recombinases (Cre, XerD, HP1 and Flp) recognize a relatively short, unique nucleic acid sequence, which serves for both recognition and recombination.1,2,4 One of the most widely used site-specific recombinases is the enzyme Cre recombinase, from the bacteriophage P1. Cre recombinase recognizes DNA at a 34 bp sequence called loxP, which consists of two 13 bp palindromic sequences flanking an 8 bp core sequence. If there are two loxP sites in the same orientation near each other (floxed), Cre recombinase can act to loop out the sequence between the two sites, leaving a single loxP site in the original DNA and a second loxP in a circular piece of DNA containing the intervening sequence. By inserting a floxed STOP cassette in the intervening sequence between the potentially toxic transgene and its promoter, one can prevent the expression of the potentially toxic gene. After establishment of a transgenic line, the STOP signal can be removed by Cre recombinase-mediated excision to activate the transgene expression.5 Thus a properly designed targeting construct containing loxP sites can be used for temporally or spatially controlled knockout or for introducing subtle mutations (for a review of the control of transgenes, see Sauer6).
The use of floxed STOP cassette has been demonstrated previously.5 The SV40 tumor antigen was rendered completely quiescent by the insertion of a synthetic STOP sequence between it and a lens-specific promoter. The resulting transgenic lines exhibited no incidence of tumor formation. On mating with a Cre recombinase-expressing transgenic mouse to generate doubly transgenic mice, recombinational activation of the tumor antigen occurred and resulted in the development of lens tumors in the offspring. The Cre/loxP system has also been used in gene deletion systems to bypass the embryonic lethalty phenotypes. The functional domain of a tumor suppressor gene was deleted in the brain using a Cre/loxP system.7,8 This approach allowed the authors to study the deletion of this gene in the brain while the conventional deletion of this gene produced a lethal phenotype.9
Many others have demonstrated that conditional gene activation or deletion can be achieved by crossing transgenic mice that have the gene of interest flanked by two loxP elements with Cre recombinase-expressing transgenic mice.8,10 In addition, conditional gene activation or deletions have also been achieved by adenoviral delivery of Cre recombinase in transgenic mice carrying the floxed gene.11 Delivery of genes by means of adenovirus-based vectors was shown to be highly efficient and applicable to a wide variety of cells types, in cell culture and in vivo in living animals. Recombination at high frequency in a range of somatic tissues was achieved in flox LacZ indicator mouse strain by the intravenous injection (1×109 PFU) of adenovirus expressing Cre recombinase (Ad-CMV-Cre).11 The liver-specific ablation of human angiotensinogen gene flanked by loxP sites in transgenic mice using an adenovirus (1.1×1011 PFU) expressing Cre recombinase (Ad-CMV-Cre) has also been reported.12
Presently in a majority of studies involving the Cre–loxP system, LacZ and fluorescent proteins (eg GFP) are being used as the reporter gene.13 The use of theses reporters requires killing the animals and ex vivo analysis for reporter gene expression. In this study, we demonstrate noninvasive imaging of Cre–loxP-mediated tissue-specific deletion using positron emission tomography (PET). The herpes simplex virus type 1 thymidine kinase (HSV1-tk), a PET reporter gene (PRG)14 that is conditionally activated following Cre–loxP-mediated deletion, is imaged in the microPET scanner (Figure 1). The HSV1-tk is one of the most widely used PET reporter gene systems. This reporter gene encodes for the viral thymidine kinase enzyme (HSV1-TK) that is capable of trapping a tracer (a radiolabeled substrate) within the cell (note: HSV1-tk refers to the gene, HSV1-TK refers to the enzyme). Acycloguanosines (eg acyclovir, ACV) and guanosines (eg ganciclovir, GCV; penciclovir, PCV) are prodrugs that are much more effectively phosphorylated by the HSV1-TK than by the mammalian TKs. When radiolabeled versions of these prodrugs are used in nonpharmacological quantities (tracer doses), they can serve as PET reporter probes. The side-chain fluorinated analog of penciclovir, 9-(4-[18F]-fluoro-3-hydroxymethyl-butyl)guanine ([18F]-FHBG),15 and 124I-labeled 2′-fluoro-2′-deoxy-1-β-d-arabinofuranosyl-5-iodouracil ([124I]-FIAU) are the most commonly used probes for imaging HSV1-tk expression.16
Figure 1.
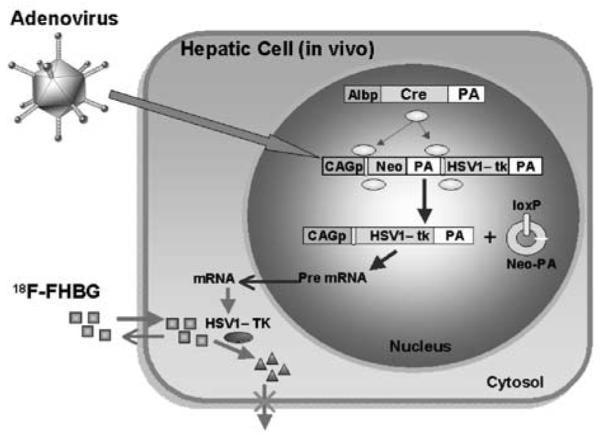
Adenovirus carrying the silent PET reporter gene delivers its genetic information (CAGp-loxP-Neo-PA-loxP-HSV1-tk-PA) into the hepatic cells. In this construct, the SV40 polyA is placed downstream of Neo and the rabbit β-globin polyA is used downstream of HSV1-tk. In the liver of transgenic mice, the albumin promoter (Albp) drives the Cre recombinase (Cre) expression. The Cre recombinase acts upon the adenoviral DNA causing excision of the Neo-PA cassette and recombination at the loxP sites leading to the HSV1-tk-mRNA formation. The expressed HSV1-TK protein acts on the reporter probe ([18F]-FHBG, □), and consequently its metabolite (Δ) is trapped within the hepatic cells leading to its detection in the microPET scanner.
The use of a PRG allows us to monitor Cre recombinase-mediated recombination noninvasively and repeatedly in the same animal. In addition, this approach may also allow us to track the in vivo expression pattern of the Cre gene, which has been shown to be expressed globally in some cases. In these cases, the inactivation of gene through Cre recombinase often gave rise to unexpected or lethal phenotypes, which complicates further analysis of the animals. We demonstrate here that the adenovirus-delivered PRG, HSV1-tk, is activated when the Cre recombinase is expressed in the liver using a mouse model. The Cre recombinase excises a stuffer DNA (neo+polyA) placed between the HSV1-tk and a CAG promoter. This PRG expression is then imaged repeatedly and noninvasively in the liver to demonstrate the efficient excision of the stuffer sequence and thus activation of the gene. This proof-of-principle experiment validates that the use of PET can enhance the studies on conditional gene modulation using the Cre–loxP system.
Results
A transcriptionally silent HSV1-tk PRG can be made active in cell cultures using the Cre–loxP system
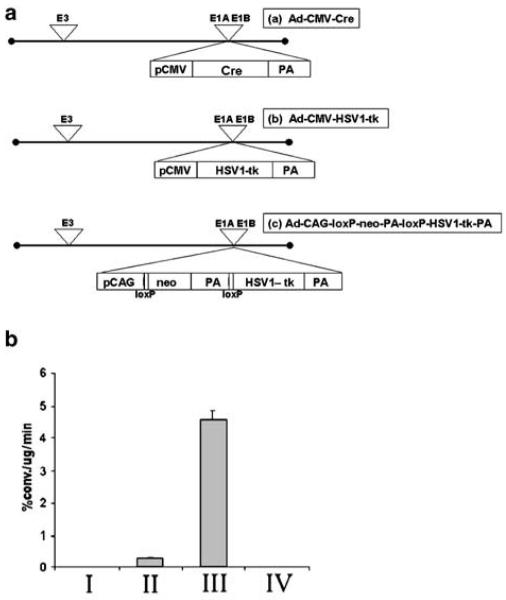
Various cell lines such as C6, 293T, HeLa, N2a and LS174T were infected with Ad-CMV-HSV1-tk only or a combination of Ad-CAG-loxP-PA-loxP-HSV1-tk and Ad-CMV-Cre (1.0×109 PFU/construct). Expression of the HSV1-tk gene was monitored by measuring TK enzyme activity. The Cre-mediated activation of HSV1-tk gene expression was achieved in all the cell lines studied (Table 1). For example, in C6 cells infected with Ad-CAG-loxP-PA-loxP-HSV1-tk and Ad-CMV-Cre, TK activity was 15 times higher than that of cells infected only with Ad-CMV-HSV1-tk (Figure 2b). The Ad-CMV-HSV1-tk-infected C6 cells had a TK activity of 0.29±0.03% conversion/μg protein/min, while the cells infected with Ad-CAG-loxP-PA-loxP-HSV1-tk and Ad-CMV-Cre combination demonstrated a TK activity of 4.56±0.24% conversion/μg protein/min (P<0.05). Similarly in the entire cell lines studied, the Cre-mediated activation of TK activity was significantly higher (P<0.05) than that induced by Ad-CMV-HSV1-tk alone (Table 1).
Table 1.
TK enzyme activity in various cell lines that were double infected with Ad-CAG-loxP-PA-loxP-HSV1-tk and Ad-CMV-Cre demonstrating Cre-mediated conditional gene activation across various cell lines
| Cell lines tested |
|||||
|---|---|---|---|---|---|
| C6 | HeLa | LS 174T | N2a | 293T | |
| Single infection with Ad-CMV-HSV1-tk | 0.29±0.03 | 0.12±0.01 | 0.02±0.01 | 0.12±0.00 | 0.26±0.12 |
| Double infection with Ad-CAG-loxP-PA- loxP-HSV1-tk and Ad-CMV-Cre | 4.56±0.24a | 0.75±0.04a | 0.12±0.00a | 3.67±0.45a | 3.35±0.26a |
Cells infected with Ad-CMV-HSV1-tk serve as positive controls. The values are mean±s.e.m.
Significantly higher (Student's t-test, P<0.05) than single virus infection.
Figure 2.
(a) Schematic diagram of the different adenoviral constructs that were used. pCMV, cytomegalovirus promoter; Cre, Cre recombinase gene; PA, polyadenylation signal; pCAG, cytomegalovirus immediate early gene 1 enhancer, chicken β-actin promoter with the β-actin intron sequence and rabbit β-globin poly-A signal. (b) Graph showing the HSV1-TK activity in the C6 cell cultures transfected with different adenoviruses: I, no adenovirus; II, Ad-CMV-HSV1-tk (1 × 109 PFU); III, double transfection of Ad-CMV-Cre (1 × 109 PFU) and Ad-CAG-loxP-PA-loxP-HSV1-tk (1 × 109 PFU); and IV, Ad-CAG-loxP-PA-loxP-HSV1-tk (1 × 109 PFU) (see also Table 1).
Infection of the cells with Ad-CAG-loxP-PA-loxP-HSV1-tk alone resulted in background levels of TK activity, which was not significantly different from noninfected cells. Thus, the TK activity is consistent with the excision of neo-PA stuffer cassette in the Ad-CAG-loxP-PA-loxP-HSV1-tk construct by Cre recombinase, which is expressed through Ad-CMV-Cre. The Ad-CAG-loxP-PA-loxP-HSV1-tk vector was shown to be readily excisable by the Cre recombinase enzyme in all the cell lines studied; therefore, we found it suitable for in vivo testing in a Cre transgenic mouse model.
Cre–loxP-mediated activation of HSV1-tk expression in murine liver can be imaged using MicroPET
To image HSV1-tk reporter gene expression in living mice, we used transgenic mice expressing Cre recombinase in the liver. In these mice, the Cre recombinase is expressed under an albumin promoter, limiting the expression of Cre recombinase to the hepatocytes. Mice expressing Cre recombinase (Cre+, n = 6) and those that did not (Cre−, n = 4) were injected with the Ad-CAG-loxP-PA-loxP-HSV1-tk vector (1.0 × 109 PFU), and expression of HSV1-tk gene was monitored using microPET. All mice were subjected to microPET scanning before, and 48 and 92 h after the tail-vein injection of virus. Hepatic accumulation of [18F]-FHBG was imaged 1 h after injection and quantified as percentage injected dose per gram of tissue (%ID/g). The expression of the PRG was evidenced by the increased hepatic accumulation of the [18F]-FHBG. In the Cre+ mice, the %ID/g liver was 0.1±0001 before viral injection and increased to 7.07±1.09 at 48 h after Ad-CAG-loxP-PA-loxP-HSV1-tk injection (Table 2; Figure 3). In three Cre recombinase-expressing mice, repeated microPET scans were performed at days 2, 4, 7 and 11 after virus injection showing that the Cre–loxP-mediated PRG activation can be followed by repeated imaging over an extended period of time. The control animals (Cre−) that did not express Cre recombinase had their %ID/g liver at less than 0.1 at all times (Table 2). The normal route of [18F]-FHBG clearance is through the gut and kidneys, and in these regions the activity was seen in all animals.
Table 2.
Data showing the thymidine kinase activity as revealed by microPET data and ex vivo analysis of liver samples
| MicroPET data (%ID/g) |
|||||
|---|---|---|---|---|---|
| Before virus inj. | After virus inj. (48 h) | First strand tk DNA copy number by QRT-PCR | First strand Cre DNA copy number by QRT-PCR | Ex vivo TK assay (% conversion/μg/min) | |
| Transgenic mice (n=6) | 0.10±0.001 | 7.07±1.09a | 33 206.67±23 352.11 | 5027.22±4268.20 | 0.50±0.2 |
| Control mice (n=4) | 0.10±0.02 | 0.20±0.09b | ND | ND | 0.01±0.001b |
The values are mean±s.e.m. ND: not detected.
Significantly higher (Student's t-test, P<0.05) than before virus injection.
Significantly lower (Student's t-test, P<0.05) than transgenic mice.
Figure 3.
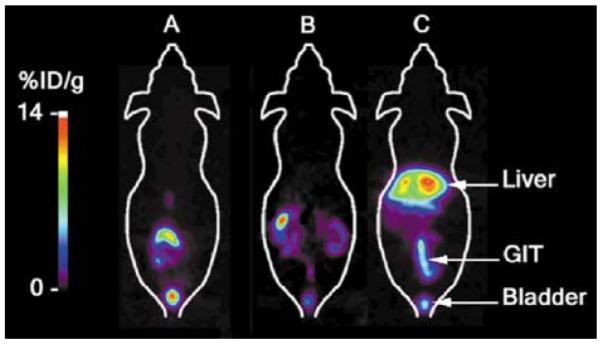
MicroPET imaging shows the HSV1-tk expression in the liver as seen by the accumulation of the tracer [18F]-FHBG in the hepatic region: (a) Cre− animal 48 h after virus injection (1 × 109 PFU of Ad-CAG-loxP-PA-loxP-HSV1-tk); (b) Cre+ animal before virus injection; and (c) 48 h after virus injection. In the pseudo-color images shown in this figure, the region of highest accumulation of [18F]-FHBG in terms of percent-injected dose per gram of tissue (%ID/g) is indicated by the white color above the red zone and the lowest is shown in purple to black color. In the images shown, the maximum accumulation of [18F]-FHBG was 14%ID/g in the liver of transgenic mice (c) infected with the above-mentioned adenovirus carrying the silent HSV1-tk reporter gene. Three-dimensional image analyses can further enhance the ability to analyze the pattern of gene expression in living animals. The activity seen around the lower abdominal area is due to the normal excretion of the tracer through the gastrointestinal tract (GIT) and renal elimination.
Measurement of HSV1-tk reporter gene expression by quantitative RT-PCR is consistent with the microPET quantitation of [18F]-FHBG accumulation
The expression of the HSV1-tk PRG in the liver was examined by quantitative RT-PCR immediately following microPET imaging. Copy number analysis of the PRG confirmed the data obtained by microPET quantitation (Table 2). The animals with increased hepatic accumulation of [18F]-FHBG (Cre+ mice) also had higher number of HSV1-tk mRNA, as compared to those with minimum [18F]-FHBG accumulation (Cre− mice). The Cre+ mice were enumerated to have 33 207±23 352 copies of first strand HSV1-tk DNA and 5027±4268 copies of first strand Cre DNA in their liver. The Cre− mice did not have detectable amount of either HSV1-tk or Cre first strand DNA (Table 2 and Figure 4a). RT-PCR from the heart and spleen tissues of Cre+ animals was negative for both Cre and tk (data not shown).
Figure 4.
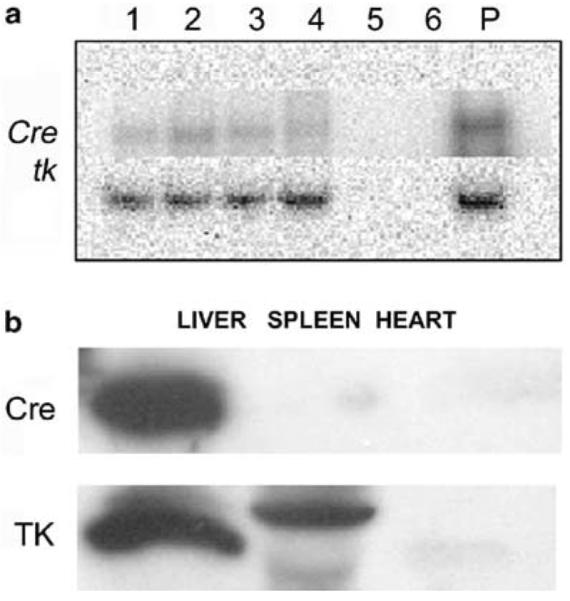
(a) Phosphor images of quantitative RT-PCR showing the expression of Cre recombinase gene (upper row) and HSV1-tk gene (lower row) in transgenic mice that express Cre recombinase (lanes 1–4) and in nontrangenic mice that do not express cre recombinase (lanes 5 and 6). Positive DNA controls for the PCR reaction are seen in lane marked as P. (b) Western blot confirmation of the HSV1-TK enzyme and Cre recombinase expression in the liver of the transgenic and nontrangenic mice (not shown) was performed by harvesting the virus-infected liver tissue following the microPET scans. Also shown is the absence of TK and Cre recombinase in the spleen and heart tissues.
Ex vivo thymidine kinase assay and Western blot analysis of liver tissues confirm the in vivo expression of HSV1-TK enzyme detected by microPET imaging
The animals that express Cre recombinase showed significantly (P<.05) higher HSV1-TK activity than the control animals (0.5±0.2 vs 0.01±0.001% conversion/μg protein/min). Western blotting analysis using anti-TK antibody showed the presence of the TK protein at the 46 kDa molecular weight region only in the Cre+ animals, demonstrating that the TK activity in the liver of these animals was due to the expression of PRG protein resulting from Cre-mediated excision and recombination of injected Ad-CAG-loxP-PA-loxP-HSV1-tk (Figure 4b). This expression of TK was also shown to colocalize to the same animals with Cre expression in their livers (Figure 4a).
Discussion
In this study, using microPET imaging, we have demonstrated that a silenced PRG (HSV1-tk) that is carried in a replication-deficient adenovirus can be activated in living mice in a tissue-specific manner using the Cre–loxP system. This proof of principle experiment reveals the potential of noninvasive imaging as a tool in studying recombinase-mediated conditional gene deletion/activation approach.
The Cre–loxP system is being widely used to create temporal and spatial control of gene deletion or gene activation in transgenic animal models created to uncover the role of a particular gene in the physiologic and/or pathologic processes. In these models, recombination in cells other than the desired target cells/tissue might occur due to inappropriate or unexpected expression of the Cre recombinase transgene, especially when driven by leaky promoters.17 Reporter genes such as LacZ10,11,18 or firefly luciferase19 have been used to monitor the level, expression pattern as well as the functionality of Cre recombinase-mediated recombination in transgenic animals. However, invasive procedures were required to analyze these reporters. We now demonstrate that by the use of a PET reporter gene it is possible to noninvasively image the expression of these reporter genes.
In our study, the location and magnitude of the expression of the Cre recombinase gene are monitored by the use of a PRG that remains silent till it is activated using the Cre–loxP system. This PRG gene activated by loxP-dependent recombination allowed us to image noninvasively the PRG expression by microPET and to identify the tissues that express the Cre recombinase. Although development of a cooled charged coupled device (CCD) camera also allows the noninvasively imaging of the expression of firefly luciferase gene in small animals,20,21 it is limited by the lack of tomographic information and full quantitation capability.
The ability to image repetitively adds another dimension to the current study. In viral vector-mediated gene transfer approaches, the limitations of long-term transgene expression include immune rejection22,23 and promoter shut off.24 In the present study, we have imaged PRG expression only up to 11 days postinjection of the adenovirus to demonstrate repetitive imaging. However, it is possible to image the reporter gene expression for longer periods of time. Previously studies have shown that the adenovirus-delivered reporter gene driven by a CMV promoter can be repetitively imaged for 88 days in the liver of Swiss-Webster mice25 for 3 months in the pituitary gland26 and for more than 150 days in the skeletal muscles of immunocompetent Swiss-Webster mice.20 Furthermore, studies have shown that combinations of the CMV promoter/enhancer with other regulatory elements such as in CAG promoter allow long-term expression.27 The Cre–loxP system further gives the advantage of gene activation using a minimal promoter that may get shut off quite earlier, but before that the gene expression driven by a potent CAG promoter would have got activated following splicing and recombination at the loxP sites. These abilities of the Cre–loxP system will allow an investigator to follow the gene expression for a long-term. For example, in experiments where gene deletion leads to cancer, the PRG can be made to activate at the same time and in the same cells where the gene of interest gets deleted. This will allow investigators to image the cancer cells and follow their serial growth, especially when the deletion of such a gene leads to invasion and metastasis.
In gene therapy studies using the HSV1-tk gene, the Cre–loxP system has been used to achieve tissue-specific and enhanced suicide gene expression. In these studies, the Cre–loxP system has been used to achieve enhanced activity of weak promoters such as thyroglobulin,17 α-fetoprotein28 and carcinoembryonic antigen.29,30 In cell cultures, Nagayama et al17 reported 5- to 10-fold higher cytotoxic effect with Cre–loxP system than that with thyroglobulin promoter alone. The enhanced activity was due to the tissue-specific Cre recombinase expression leading to the excision and recombination of loxP sites leading to the expression of suicide gene driven by the strong CAG promoter. This method of achieving enhanced activity was ideally achieved using the Cre–loxP system in cell cultures. It should be noted that in the present study in cell cultures the method of double infection involving the Cre–loxP system resulted in many fold higher level of TK activity than single infection with Ad-CMV-HSV1-tk. It should be considered that a limited number of induced Cre recombinase effectively processes a large number of molecules on the target adenoviral genome.31,32 Besides the signal amplification by the Cre–loxP system, the increased activity could also be due to the use of more potent CAG promoter in the Cre–loxP system that we used33,34 while the CMV promoter was used in single infections with Ad-CMV-HSV1-tk. Since our primary aim of this study was to demonstrate conditional activation of a PRG, we did not attempt to compare the two promoters directly. However, we have previously demonstrated the results of Ad-CMV-HSV1-tk and PET imaging in living animals.35
Goto et al29 showed tumor suppression in four of 10 mice infected with Ad-CEA-Cre and Ad-lox-TK followed by ganciclovir treatment. Sakai et al28 showed in cultured cells, 60- to 300-fold higher expression of HSV1-TK enzymatic activity and ganciclovir sensitivity through the Cre–loxP system than with the α-fetoprotein promotor alone. In these studies, two adenoviruses, one that carries the Cre recombinase gene driven by a tissue-specific promoter and another that has the floxed HSV1-tk gene driven by a strong promoter such as the CAG promoter, were used. However, they reported no significant antitumor effect for subcutaneous tumor on athymic mice. The lack of antitumor effect in mice could be explained by insufficient simultaneous infection of two viruses in the same cell of the tumor. In our studies, injection of two viruses (Ad-CMV-Cre and Ad-CAG-loxP-PA-loxP-HSV1-tk; 1 × 109 PFU each construct) in nude mice did not lead to detectable levels of TK activity (data not shown).
A single viral construct delivery to a liver-specific Cre transgenic model proved to be a good model in our system. In our experiments, we detected HSV1-TK activity by microPET imaging only in animals that carry and express the Cre recombinase gene and that were tail-vein injected with the adenovirus. In these mice, the HSV1-tk gene carried by the adenovirus was expressed due to the Cre recombinase-mediated activation of this PRG. The expression levels of this gene, confirmed by the quantitative RT-PCR, showed significantly higher copy numbers of HSV1-tk mRNA only in animals that also showed TK activity by microPET imaging. Animals that did not express Cre recombinase but injected with the adenovirus showed no detectable HSV1-TK activity by PET imaging, and their QRT-PCR also showed no detectable levels of HSV1-tk mRNA copy numbers. Injection of a lower titer of the virus might have resulted in lower levels of gene expression. However, we did not explore this aspect in the current study as we have previously observed that the dose of adenovirus injected does not correlate well with the level of reporter gene expression in the liver.35 This could be due to factors such as hydration, extravasation, etc, that are likely to influence the amount of virus that will be available at the target site(s) (eg liver). Future studies will need to correlate levels of viral dose injected, cre expression at target site and HSV1-tk reporter gene expression at the target site.
A possible way to overcome the limitation in using two viral constructs for gene therapy applications is to construct a single adenovirus bearing both the Cre–loxP system and the therapeutic gene in the genome. However, in this case, it is essential to completely suppress the activity of Cre recombinase expression when the adenovirus is being grown. If not the stuffer region will be excised and the adenovirus will lose its tissue/tumor specificity while it is being grown. To overcome the problem associated with the adenoviral-based single vector system, a plasmid-based single vector system has been studied in cell culture.36 These modifications prevented the translation of Cre recombinase in bacteria while doing amplification but allowed its translation in eucaryotic cells. However, the offset with this type of vector is its delivery in living animals. We and others have previously reported that this type of vector can be delivered into living animals using liposomes37 or by the use of polyethylenimine-poly-ethylene glycol (PEI-PEG) formulations.38 In our study, we used a single viral vector delivery system in vivo in a transgenic mouse model bypassing the problem of coinfection efficiency associated with two viral vector system. The use of a single viral vector containing both the Cre recombinase and loxP recombination sites for the delivery of tissue specific gene is currently being considered for further investigation in our laboratory.
The tail-vein injection of the adenovirus was ideal for the present study as 90–95% of the intravenously injected adenovirus infects the liver. However, to apply the use of PRG in other Cre transgenic animals, such as those that express Cre recombinase in heart or prostate, the adenovirus injection via tail vein will likely not be very useful. In such transgenic animal models or in tumor models, either direct injection of the adenovirus at the specific site or redirected adenoviruses that use various molecular bridges such as peptide-targeting ligands,39 bispecific antibodies40 or directly biotinylated adenovirus vectors41 are likely to be much useful. Alternatively, different gene delivery modalities such as liposomes37 or PEI-PEG formulations38 may also be useful. Furthermore, in transgenic animal models in which gene deletion leads to cancer, the PRG can be made to activate at the same time and in the same cells where the gene of interest gets deleted. This will allow investigators to image the cancer cells and follow their serial growth, especially when the deletion of such a gene leads to invasion and metastasis.
Tjuvajev et al16 have shown the 124I-FIAU, when used as a probe for imaging HSV1-tk expression, to be more efficient than [18F]-FHBG or 18F-labeled 9-[3-fluoro-1-hydroxy-2-propoxymethyl]guanine ([18F]-FHPG), with greater sensitivity and contrast as well as lower levels of abdominal background radioactivity at 2 and 24 h. However, in our studies, we used [18F]-FHBG as the reporter probe for imaging HSV1-tk expression, as one of our goals is to do repeated imaging to see if the degree of HSV1-tk expression goes down over time due to the immune activity against the adenovirus in the mice. This will not be possible if a probe with 124I label is used due to its longer half-life of ~4 days mandating a minimum waiting period of 10–12 days prior to repeat imaging. Future studies may also use HSV1-sr39tk and [18F]-FHBG, which we have demonstrated are likely to be more sensitive than HSV1-tk/FIAU.42
Reporter genes such as LacZ10,11,18 or firefly luciferase19 have been used to monitor the level and functionality of Cre recombinase-mediated recombination in transgenic animals. However, in the above-mentioned studies, invasive procedures were used to study these reporter systems where the animals were killed to know the extent of gene expression. However, we now demonstrate that by the use of a PET reporter gene it is possible to image noninvasively the expression of such reporter genes. In the study mentioned above,19 the authors have used invasive methods to study the expression of firefly luciferase gene. However, it is now possible to image noninvasively the expression of firefly luciferase gene in small animals20,21 using a cooled CCD camera. The CCD camera may offer a cheap alternative to microPET for noninvasive imaging but with the limitation of lack of tomographic information. Other reporters under development in our lab, such as the synthetic Renilla luciferase (hRluc), have been shown to be highly sensitive, and their expression could be detected in only a few hundred cells in a living mouse.43 This optical reporter gene could be well suited to monitor such low levels of transcriptional read-throughs with a cooled CCD camera. More recently, a novel reporter vector was constructed encoding a fusion protein comprising a mutant HSV1-tk (HSV1-sr39tk) and Rluc joined by a 20 amino-acid long spacer sequence.44 This will allow multimodality imaging of the reporter gene expression in both a PET scanner and cooled CCD camera.
In addition to the cre/loxP strategy that can be used to achieve tissue specificity with higher levels of transgene expression, a two-step transcriptional amplification/activation (TSTA) strategy has also been validated.45,46 In the TSTA strategy, a potent transcriptional activator, driven by a tissue-specific promoter, activates a GAL4-VP16 fusion protein, which, in turn, drives reporter gene expression.47,48 It is important that any reporter gene-imaging approach not significantly perturb the cells/animal models being studied. However, there is potential for the GAL4-VP16 system to be toxic to cells.49,50 In zebrafish, high levels of injected GAL4-VP16 were deleterious to development.50 It may be the case that levels of GAL4-VP16 will need to be modulated to strike a balance between amplification and any potential toxicity. Similar toxicity issues do exist for the Cre–loxP system also. Loonstra et al51 have shown that Cre recombinase expression in cultured mammalian cells may result in a markedly reduced proliferation and also numerous chromosomal aberrations and an increased number of sister chromatid exchanges. However, they also demonstrated that the toxicity is dependent on the level of Cre recombinase activity. Prolonged low levels of Cre recombinase activity permit recombination without concomitant toxicity.51 The TSTA strategy has the advantage that reporter gene expression can be modulated, whereas in the Cre–loxP strategy once cre recombinase cuts specific sites this cannot be reversed. The Cre–loxP system may have an advantage over the TSTA system, as the Cre recombinase being an enzyme will require only small quantities for an effective conditional gene activation/amplification process. Cre recombinase once made will persist in cells and can make repeated recombinations in vectors having the appropriate loxP sites. Because of this property, it might be possible to operate this system at very low levels of Cre recombinase expression. Future studies will need to focus on differential levels of Cre recombinase expression to evaluate the sensitivity of the PRG-based Cre–loxP system.
By microPET imaging, we have now shown that a PRG that is conditionally activated by the Cre–loxP system can be repetitively imaged in an animal model. Appropriate use of a reporter gene that can be imaged noninvasively will provide investigators with a powerful tool to monitor the location, magnitude and duration of Cre-mediated conditional gene activation/deletion in transgenic animals or in animal tumor models.
Materials and methods
Cell lines
The C6 rat glioma cells were kindly provided by Margaret Black (Washington State University, Pullman) and were grown in deficient DMEM, supplemented with 5% FBS and 1% penicillin/streptomycin/l-glutamine. The N2a cells were obtained from VP Mauro (Scripps Research Institute, La Jolla, CA, USA) and were grown in DMEM (high glucose) supplemented with 10% FBS and 1% penicillin/streptomycin. HeLa cells (ATTC – # CCL2) were grown in MEM with 2 mm l-glutamine and 0.1 mm nonessential amino acids, and 1.0 mm sodium pyruvate, 1% penicllin/streptomycin and 10% fetal bovine serum. LS 174T cells (ATCC – # CL188) were grown in DMEM/F12 Ham media with 5% FBS and 1% penicillin/streptomycin. All cell lines were maintained by incubating at 37°C with 5% CO2. 293 cells (ATCC – # CRL1573) were used for growth and titration of adenovirus vectors (see below). C6 cells were used to test the Cre–loxP-mediated recombination to activate the PRG in cell cultures. The confluent C6 cells grown in 10 cm Petri dishes were infected with 1 × 109 PFU of the adenovirus and assayed 48 h later for HSV1-TK enzyme. Similarly, the system was also studied in other cell lines such as 293T, HeLa, N2a and LS-174T to test the efficiency of the system across different cell types.
Adenoviral vectors
We used E1/E3-deleted recombinant adenoviral vector to construct our delivery vectors. All vectors were constructed using the COS-TPC method.52 All the adenoviruses used in this study were propagated, purified and stored at −70°C, as described.53 Titers of viral preparations were determined by plaque assays using 293 cells.54 In order to test the excision efficiency of the STOP cassette by Cre recombinase, we generated three testing constructs: Ad-CMV-HSV1-tk, Ad-CMV-Cre and Ad-CAG-loxP-PA-loxP-HSV1-tk (Figure 2a). The Ad-CMV-HSV1-tk construct contains the HSV1-tk gene driven by a robust promoter, thus is used as a control construct for HSV1-tk expression. The Ad-CMV-Cre construct was used to express the Cre recombinase in the in vitro system to test the efficiency of the Ad-CAG-loxP-PA-loxP-HSV1-tk, which was then used in vivo. The Ad-CAG-loxP-PA-loxP-HSV1-tk is the construct that contains the inactivated HSV1-tk gene. The HSV1-tk gene is separated from the CAG promoter by a stuffer sequence containing neo/SV40 polyA. The stuffer sequence is flanked by two loxP sequences allowing cleavage of the stuffer gene by Cre recombinase. The PolyA signal (PA) stops the gene expression at the transcriptional level, thus no neo protein would be made. In this system, the neo mRNA is produced before Cre recombinase-mediated recombination and HSV1-tk mRNA is produced after Cre recombinase-mediated recombination wherein the floxed neo gene is excised.
Transgenic mice
Transgenic mice that express Cre recombinase driven by the albumin promoter (Alb-Cre) were obtained from the Jackson Laboratory (Maine, USA). The sense and anti-sense primers of Cre recombinase (sense 5′ CGT ATA GCC GAA ATT GCC AG 3′ and antisense 5′ CAA AAC AGG TAG TTA TTC GG 3′) were used for genotyping by setting 30 PCR cylces with denaturation at 95°C for 1 min, annealing at 53°C for 1 min and extension at 72°C for 1 min, followed by final extension for 5 min. The expected PCR product of 190 base pairs was confirmed by running 1% agarose gel with suitable marker. DNA extracted from Cre− mice served as negative controls.
MicroPET imaging
Mice were imaged using the microPET small animal scanner.55,56 Mice were injected via tail vein with ~200 μCi of [18F]-FHBG (specific activity 5–10 Ci/μmol) synthesized as described previously.57 After an hour uptake time had elapsed, mice were anesthetized using a mixture of ketamine (80 mg/kg) and xylazine (10 mg/kg) by intraperitoneal injection and placed in a supine position, and imaged using the microPET scanner with the long axis of the mouse parallel to the long axis of the scanner. Acquisition time was 28 min (4 min per bed position; seven bed positions), and images were reconstructed using a filtered back projection (FBP) reconstruction algorithm. All mice were imaged in the microPET scanner before the tail-vein injection of 1×109 PFU of the adenovirus with Ad-CAG-loxP-PA-loxP-HSV1-tk to serve as their own controls. These animals were imaged again in the microPET scanner 48 and 92 h after adenovirus injection. Three mice were scanned for up to 11 days after virus administration. Following the scan, mice were euthanized and the tissues were processed for thymidine kinase enzyme assay, quantitative RT-PCR and western blotting. All animal handling procedures were performed in accordance with UCLA Animal Research Committee guidelines.
MicroPET data analysis
Quantitation of microPET images was performed using the Crump Institute Integrated Imaging Software Package (CRIIISP) (Crump Institute for Molecular Imaging, UCLA). From the 3-D reconstruction, several planes (out of the total 64) encompassing the liver were selected in the coronal orientation and averaged. Region of interest (ROI) were drawn centered on the peak of the activity profile. Near equal-sized ROI were drawn on all occasions. ROI counts/pixel/min were converted to counts/cm3/min using a calibration factor obtained from scanning a cylinder containing a known amount of 18F activity. The ROI counts/ml/min were converted to counts/g/min (assuming a tissue density of 1 g/ml) and divided by the injected dose to obtain an image ROI-derived [18F]-FHBG percentage injected dose per gram of liver (%ID/g). No corrections for partial volume or attenuation were performed. The effects of ROI positioning were determined by averaging at least three ROI and assessing the variability across regions.
Thymidine kinase enzyme assays
For HSV1-TK assays, cultured cells or liver tissues were homogenized and enzyme activity determined as described.58 Briefly, the radioisotope 8-[3H]penciclovir (8-[3H]PCV) (11 Ci/mmol) was used as the substrate (Moravek Biochemicals, La Brea, CA, USA). The results were expressed as percent conversion of 8-[3H]PCV (% conversion/mg protein/min)=dpm/μg protein/min of cell or tissue extract)/(dpm of control sample)×100.
Western blot analysis
The liver, spleen and heart tissues were dissected from the euthanized animals and lysed mechanically in a buffer containing 10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mm DTT with 20% glycerol and 0.1 mm PMSF. The samples were centrifuged at 4°C, 10 000 rpm for 5 min. Protein was determined using a Biorad protein assay reagent (Hercules, CA, USA), and 50 μg of protein from each sample was mixed with two volumes of sample buffer and boiled for 5 min. Denatured samples were electrophoresed in 12% acrylamide gel and transferred to PVDF membrane using Hoefer semidry blotting apparatus. The membrane was immediately transferred to TBS (100 mm Tris pH 7.5, 150 mm NaCl, 0.05% Tween 20) containing 3% milk powder and blocked overnight with proper mixing. The membrane was then incubated with primary antibody (polyclonal rabbit anti-TK antibody, kind gift from Margaret Black, Washington University; or with anti-Cre antibody; Novagen) for 1 h at room temperature with proper shaking. The washed membrane was incubated for 30 min with goat anti-rabbit IgG-HRP conjugate (Promega) at room temperature. Immunochemical detection was carried out using the substrates from ECL kit (Amersham) by exposing the film for 30 s.
Quantitative RT-PCR
Total RNA was prepared from the liver tissue using the RNeasy mini-kit (Qiagen). DNA contamination was removed by treating the RNA samples with Rnase free Dnase (Amerhsam). Dnase was removed by performing the RNA clean-up procedure using the RNeasy mini-kit (Qiagen). To prepare first strand cDNA, 1 μg of total RNA was incubated in 20 μl of reaction mix containing 2 μl of first strand buffer, 1 μl dNTP mix (10 mm each), 1 μl 100 mm DTT, 1 μl powerscript reverse transcriptase (Clontech) and 0.1 mm random primers (Promega). The HSV1-tk and the Cre recombinase genes were amplified from the cDNA by PCR with synthesized primers (HSV1-tk sense 5′ GTA ATG ACA AGC GCC CAG AT 3′ and anti-sense 5′ATG CTG CCC ATA AGG TAT CG 3′; Cre sense 5′ CGT ATA GCC GAA ATT GCC AG 3′ and anti-sense 5′ CAA AAC AGG TAG TTA TTC GG 3′). In all, 5 μl of the first strand DNA and known copy numbers of plasmid DNA containing HSV1-tk gene were used for PCR amplification with 100 pmol of each sense and antisense primers in a solution containing 20 mm Tris-HCl (pH 8.0), 50 mm KCl, 1.5 mm MgCl2, 1.5 U DNA polymerase (Promega), 0.25 mm dATP, 0.25 mm dGTP, 0.25 mm dTTP, 0.25 μM dCTP and 5 μCi [α-32P]dCTP (Amersham). For amplifying the HSV1-tk gene, 30 PCR cycles were performed with denaturation at 95°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 1 min, followed by final extension for 5 min. For the amplification of Cre gene, the same PCR conditions with 53°C as annealing temperature was used. After PCR, samples (5 μl) were subjected to acrylamide gel electrophoresis (6% gel), and radioactive bands were detected and quantified using a Fuji Image Analyzer Bas 2000. DNA copy numbers in the standards were calculated using the formula 1 μg of 1000 bp DNA = 9.1×1011 copies. The unknown copy numbers of the samples were deduced by comparing the band intensity of known copy numbers of the plasmid DNA with the band intensities seen in the experimental samples by using the MacBAS Ver 2.4 software.
Statistical analysis
All data were subjected to statistical analysis using Excel software (Microsoft, 2000) package. Two-tailed Student's t-test was used to determine the significant differences between the groups. Differences were considered significant at P<0.05. Data are given as mean±s.e.m.
Acknowledgements
We thank Xiaoman Lewis, Judy Edwards and Waldmar Ladno (Crump Institute for Molecular Imaging) for technical assistance. We would also like to thank Dr Lily Wu and Jamie Matherly, UCLA for their help with adenoviral work. This work was supported by funding from Department of Energy contract DE-FC03-87ER60615 (SSG), NIH RO1 CA82214-01 (SSG) and SAIRP R24 CA92865 (SSG).
References
- 1.Grainge I, Jayaram M. The integrase family of recombinase: organization and function of the active site. Mol Microbiol. 1999;33:449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- 2.Voziyanov Y, Pathania S, Jayaram M. A general model for site-specific recombination by the integrase family recombinases. Nucleic Acids Res. 1999;27:930–941. doi: 10.1093/nar/27.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoess RH, Ziese M, Sternberg N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci USA. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo F, Gopaul DN, Van Duyne GD. Asymmetric DNA bending in the Cre–loxP site-specific recombination synapse. Proc Natl Acad Sci USA. 1999;96:7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakso M, et al. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci USA. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 7.Groszer M, et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 8.Lesche R, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 9.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 10.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 11.Akagi K, et al. Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res. 1997;25:1766–1773. doi: 10.1093/nar/25.9.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stec DE, et al. Efficient liver-specific deletion of a floxed human angiotensinogen transgene by adenoviral delivery of Cre recombinase in vivo. J Biol Chem. 1999;274:21285–21290. doi: 10.1074/jbc.274.30.21285. [DOI] [PubMed] [Google Scholar]
- 13.Kawamoto S, et al. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470:263–268. doi: 10.1016/s0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- 14.Gambhir SS, et al. Imaging transgene expression with radionuclide imaging technologies. Neoplasia. 2000;2:118–138. doi: 10.1038/sj.neo.7900083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer M, et al. 8-[18F]fluoropenciclovir: an improved reporter probe for imaging HSV1-tk reporter gene expression in vivo using PET. J Nucl Med. 2001;42:96–105. Comment in: J Nucl. Med. 2001 Jan;42(1):106–9 UI: 21038008. [PubMed] [Google Scholar]
- 16.Tjuvajev JG, et al. Comparison of radiolabeled nucleoside probes (FIAU, FHBG, and FHPG) for PET imaging of HSV1-tk gene expression. J Nucl Med. 2002;43:1072–1083. [PubMed] [Google Scholar]
- 17.Nagayama Y, et al. Enhanced efficacy of transcriptionally targeted suicide gene/prodrug therapy for thyroid carcinoma with the Cre-loxP system. Cancer Res. 1999;59:3049–3052. [PubMed] [Google Scholar]
- 18.Wagner KU, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anton M, Graham FL. Site-specific recombination mediated by an adenovirus vector expressing the Cre recombinase protein: a molecular switch for control of gene expression. J Virol. 1995;69:4600–4606. doi: 10.1128/jvi.69.8.4600-4606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JC, Sundaresan G, Iyer M, Gambhir SS. Noninvasive optical imaging of firefly luciferase reporter gene expression in skeletal muscles of living mice. Mol Ther. 2001;4:297–306. doi: 10.1006/mthe.2001.0460. [DOI] [PubMed] [Google Scholar]
- 21.Contag CH, et al. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 22.Tripathy SK, Black HB, Goldwasser E, Leiden JM. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, et al. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loser P, Jennings GS, Strauss M, Sandig V. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NFkappaB. J Virol. 1998;72:180–190. doi: 10.1128/jvi.72.1.180-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Q, et al. Monitoring adenoviral DNA delivery, using a mutant herpes simplex virus type 1 thymidine kinase gene as a PET reporter gene. Gene Therapy. 2002;9:1659–1666. doi: 10.1038/sj.gt.3301899. [DOI] [PubMed] [Google Scholar]
- 26.Southgate TD, et al. Long-term transgene expression within the anterior pituitary gland in situ: impact on circulating hormone levels, cellular and antibody-mediated immune responses. Endocrinology. 2001;142:464–476. doi: 10.1210/endo.142.1.7898. [DOI] [PubMed] [Google Scholar]
- 27.Kiwaki K, et al. Correction of ornithine transcarbamylase deficiency in adult spf(ash) mice and in OTC-deficient human hepatocytes with recombinant adenoviruses bearing the CAG promoter. Hum Gene Ther. 1996;7:821–830. doi: 10.1089/hum.1996.7.7-821. [DOI] [PubMed] [Google Scholar]
- 28.Sakai Y, et al. Gene therapy for hepatocellular carcinoma using two recombinant adenovirus vectors with alpha-fetoprotein promoter and Cre/lox P system. J Virol Methods. 2001;92:5–17. doi: 10.1016/s0166-0934(00)00240-8. [DOI] [PubMed] [Google Scholar]
- 29.Goto H, et al. Gene therapy utilizing the Cre/loxP system selectively suppresses tumor growth of disseminated carcinoembryonic antigen-producing cancer cells. Int J Cancer. 2001;94:414–419. doi: 10.1002/ijc.1474. [DOI] [PubMed] [Google Scholar]
- 30.Kijima T, et al. Application of the Cre recombinase/loxP system further enhances antitumor effects in cell type-specific gene therapy against carcinoembryonic antigen-producing cancer. Cancer Res. 1999;59:4906–4911. [PubMed] [Google Scholar]
- 31.Kanegae Y, et al. Efficient gene activation system on mammalian cell chromosomes using recombinant adenovirus producing Cre recombinase. Gene. 1996;181:207–212. doi: 10.1016/s0378-1119(96)00516-1. [DOI] [PubMed] [Google Scholar]
- 32.Kanegae Y, et al. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 1995;23:3816–3821. doi: 10.1093/nar/23.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 34.Xu L, et al. CMV-beta-actin promoter directs higher expression from an adeno-associated viral vector in the liver than the cytomegalovirus or elongation factor 1 alpha promoter and results in therapeutic levels of human factor X in mice. Hum Gene Ther. 2001;12:563–573. doi: 10.1089/104303401300042500. [DOI] [PubMed] [Google Scholar]
- 35.Gambhir SS, et al. Imaging adenoviral-directed reporter gene expression in living animals with positron emission tomography. Proc Natl Acad Sci USA. 1999;96:2333–2338. doi: 10.1073/pnas.96.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaczmarczyk SJ, Green JE. A single vector containing modified cre recombinase and LOX recombination sequences for inducible tissue-specific amplification of gene expression. Nucleic Acids Res. 2001;29:E56–56. doi: 10.1093/nar/29.12.e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer M, Berenji M, Templeton NS, Gambhir SS. Noninvasive imaging of cationic lipid-mediated delivery of optical and PET reporter genes in living mice. Mol Ther. 2002;6:555–562. doi: 10.1006/mthe.2002.0700. [DOI] [PubMed] [Google Scholar]
- 38.Hildebrandt IJ, Iyer M, Wagner E, Gambhir SS. Optical imaging of transferrin targeted PEI/DNA complexes in living subjects. Gene Therapy. 2003;10:758–764. doi: 10.1038/sj.gt.3301939. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, et al. Targeting adenoviral vectors by using the extracellular domain of the coxsackie-adenovirus receptor: improved potency via trimerization. J Virol. 2002;76:1892–1903. doi: 10.1128/JVI.76.4.1892-1903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nettelbeck DM, et al. Targeting of adenovirus to endothelial cells by a bispecific single-chain diabody directed against the adenovirus fiber knob domain and human endoglin (CD105) Mol Ther. 2001;3:882–891. doi: 10.1006/mthe.2001.0342. [DOI] [PubMed] [Google Scholar]
- 41.Smith JS, et al. Redirected infection of directly biotinylated recombinant adenovirus vectors through cell surface receptors and antigens. Proc Natl Acad Sci USA. 1999;96:8855–8860. doi: 10.1073/pnas.96.16.8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyer M, et al. Comparison of FPCV, FHBG, and FIAU as reporter probes for imaging Herpes Simplex Virus Type 1 thymidine kinase reporter gene expression. J Nucl Med. 2000;41:80P–81P. [PubMed] [Google Scholar]
- 43.Bhaumik S, Gambhir SS. Simultaneous imaging of the expression of two bioluminescent reporter genes in living mice. Mol Ther. 2002;5:S422. [Google Scholar]
- 44.Ray P, Wu AM, Gambhir SS. Optical bioluminescence and positron emission tomography imaging of a novel fusion reporter gene in tumor xenografts of living mice. Cancer Res. 2003;63:1160–1165. [PubMed] [Google Scholar]
- 45.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 46.Emami KH, Carey M. A synergistic increase in potency of a multimerized VP16 transcriptional activation domain. EMBO J. 1992;11:5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iyer M, et al. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci USA. 2001;98:14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, et al. Molecular engineering of a two-step transcription amplification (TSTA) system for transgene delivery in prostate cancer. Mol Ther. 2002;5:223–232. doi: 10.1006/mthe.2002.0551. [DOI] [PubMed] [Google Scholar]
- 49.Braselmann S, Graninger P, Busslinger M. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. Proc Natl Acad Sci USA. 1993;90:1657–1661. doi: 10.1073/pnas.90.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- 51.Loonstra A, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyake S, et al. Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc Natl Acad Sci USA. 1996;93:1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenfeld MA, et al. Adenovirus-mediated transfer of a recombinant alpha 1-antitrypsin gene to the lung epithelium in vivo. Science. 1991;252:431–434. doi: 10.1126/science.2017680. [DOI] [PubMed] [Google Scholar]
- 54.Kolls J, Peppel K, Silva M, Beutler B. Prolonged and effective blockade of tumor necrosis factor activity through adenovirus-mediated gene transfer. Proc Natl Acad Sci USA. 1994;91:215–219. doi: 10.1073/pnas.91.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cherry SR, Gambhir SS. Use of positron emission tomography in animal research. ILAR J. 2001;42:219–232. doi: 10.1093/ilar.42.3.219. [DOI] [PubMed] [Google Scholar]
- 56.Cherry SR, et al. MicroPET: a high resolution PET scanner for imaging small animals. IEEE Trans Nucl Sci. 1997;44:1161–1166. [Google Scholar]
- 57.Yaghoubi S, et al. Human pharmacokinetic and dosimetry studies of [(18)F]FHBG: a reporter probe for imaging herpes simplex virus type-1 thymidine kinase reporter gene expression. J Nucl Med. 2001;42:1225–1234. Comment in: J Nucl Med. 2001 Aug;42(8):1235–7 UI: 21376327. [PubMed] [Google Scholar]
- 58.Gambhir SS, et al. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc Natl Acad Sci USA. 2000;97:2785–2790. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]