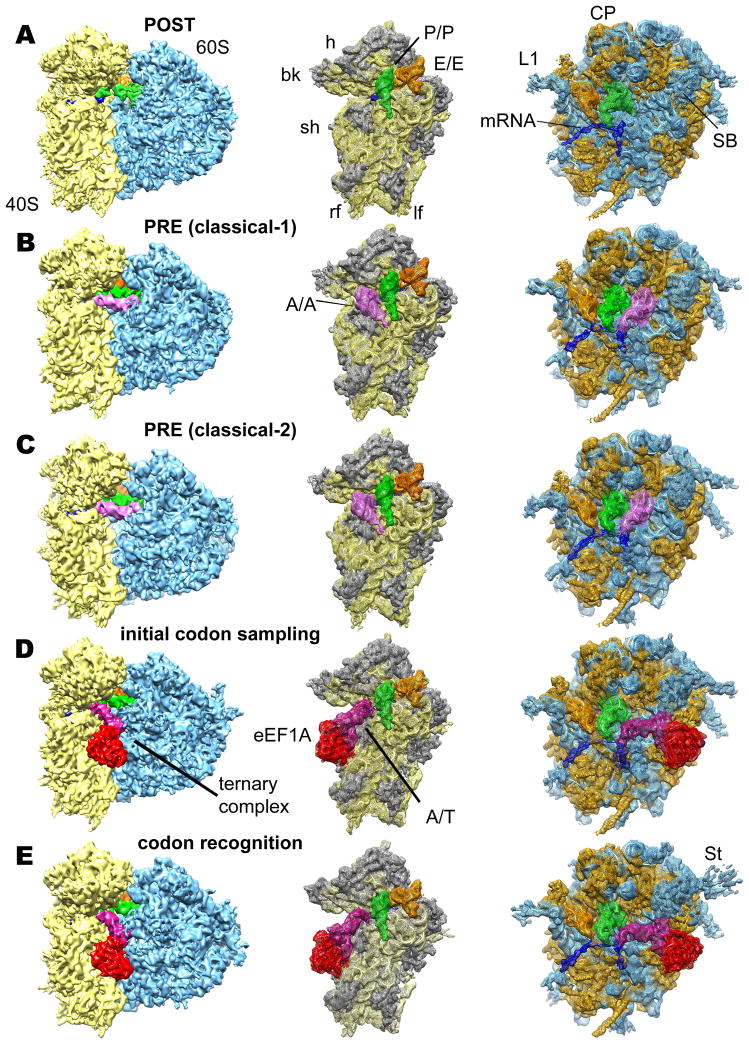
Figure 1. Reconstruction of eukaryotic 80S POST (A), PRE (classical-1) (B), PRE (classical-2) (C), codon sampling (D) and codon recognition / GTPase activation (E) complexes from rabbit liver.
Left panels: Overall view of the cryo-EM reconstructions, displaying the 60S subunit (blue), 40S subunit (yellow), A-tRNA (pink), P-tRNA (green), E-site tRNA (orange), A/T-tRNA (dark violet), eEF1A (red) and mRNA (blue). Middle and right panels: Individual mesh representation of the subunit maps with docked models, showing the ribosomal ligands relative to the 40S subunit (ribosomal RNA yellow and S proteins gray) and the 60S subunit (rRNA blue and L proteins orange), respectively. Landmarks are denoted for 40S: beak (bk), left foot (lf), right foot (rf), head (h), shoulder (sh) and 60S: central protuberance (CP), L1 stalk (L1), stalk base (SB) and stalk (St).
See also
Figure S1. Heterogeneity in the 80S ribosome POST complexes from rabbit liverand resolution curves.
Table S1. Summary of the modelled proteins in the rabbit ribosome.