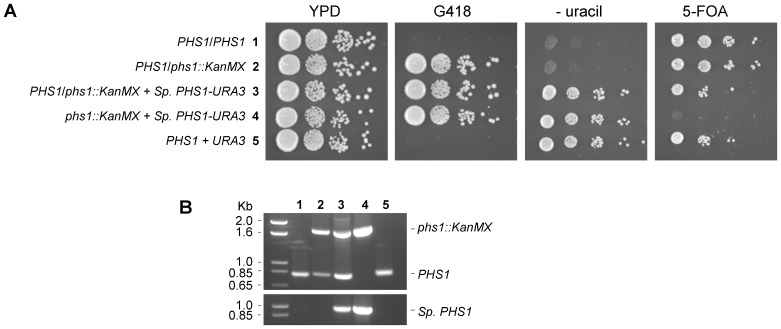
Figure 3. Sporobolomyces PHS1 complements loss-of-function of the S. cerevisiae phs1 homolog.
Five strains (numbered 1–5) were cultured on different media (A) and the presence or absence of the native PHS1 or the Sporobolomyces PHS1 gene in S. cerevisiae tested by PCR (B). (A) Ten fold serial dilutions of yeast strains. YPD allows equal growth of the strains. G418 selects for strains carrying a phs1::KanMX allele. The YNB – uracil selects for strains carrying a plasmid with the URA3 selectable marker. 5-FOA is used to counter-select or “cure” plasmids containing the URA3 marker from a ura3 mutant strain background. Strain 1 is a diploid with both copies of PHS1. Strain 2 is a heterozygote strain with one wild type and one mutated copy of the PHS1 gene. Strain 3 is strain 2 expressing the Sporobolomyces PHS1 gene from a 2 µ plasmid. Strain 4 is a haploid progeny from meiosis of strain 3. Strain 5 is a control haploid strain with the 2 µ plasmid. (B) The top panel shows PCR of the wild type allele of S. cerevisiae PHS1 (near the 0.85 kb ladder marker) or the replacement allele phs1::KanMX (near to 1.6 kb ladder marker). The bottom panel is amplification of the Sporobolomyces PHS1 gene from the same strains.