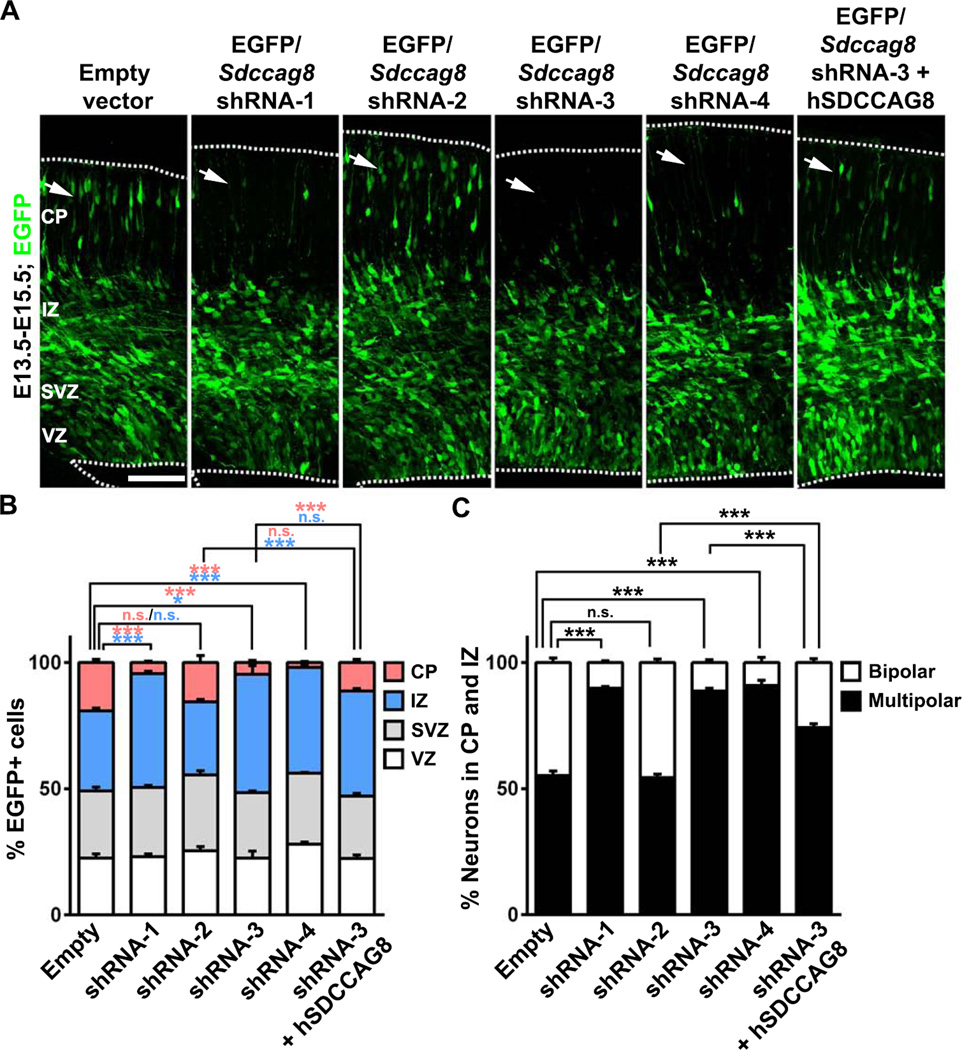
Figure 2. Depletion of SDCCAG8 disrupts neuronal distribution in the developing cortex.
(A) Representative images of E15.5 cortices electroporated with EGFP/empty vector control, Sdccag8 shRNA1-4, or Sdccag8 shRNA-3 together with shRNA-resistant human SDCCAG8 (hSDCCAG8) at E13.5. Note a lack of transfected cells expressing EGFP (green) in the CP (arrows) in cortices expressing Sdccag8 shRNA-1, −3 and −4, and the partial rescue of this defect by co-expression of shRNA-resistant hSDCCAG8. Broken lines indicate the pial surface and the VZ surface. Scale bar: 100 µm (B) Quantification of the percentage of EGFP-expressing (EGFP+) cells in different regions of the developing cortex. Data are presented as mean ± s.e.m. (Empty vector, n = 926 cells; shRNA-1, n = 791 cells; shRNA-2, n = 989 cells; shRNA-3, n = 1,150 cells; shRNA-4, n = 1,444 cells; shRNA-3 + hSDCCAG8, n = 1,305 cells; 5 brains for each condition). ***, p<0.001; *, p<0.05; n.s., not significant. (C) Quantification of the percentage of EGFP+ cells in the IZ and the CP with a bipolar or multipolar morphology. Data are presented as mean ± s.e.m. (Empty vector, n = 1,283 cells; shRNA-1, n = 773 cells; shRNA-2, n = 1,267 cells; shRNA-3, n = 1,071 cells; shRNA-4, n = 867 cells; shRNA-3 + hSDCCAG8, n = 765 cells; 5 brains for each condition). ***, p<0.001. n.s., not significant.