Figure 3.
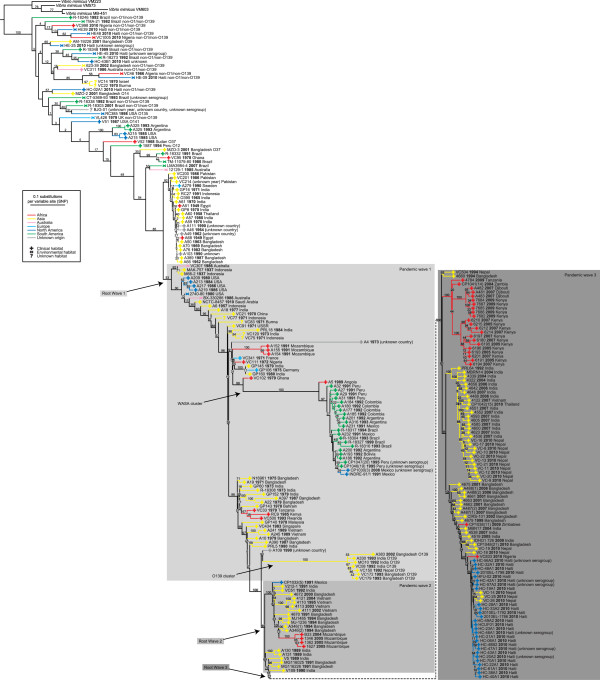
Phylogenomic tree of V. cholerae genomes. A phylogenomic tree based on genome-wide marker SNPs illustrates the breadth of 274 V. cholerae genomes included in this study. Four complete V. mimicus genomes were included as an outgroup. Branch lengths indicate the number of substitutions per SNP site. Several clusters mentioned in the text are shown. Branches are colored by the continent where the strains were isolated. The isolation source (habitat) of the strains is indicated. All strains belong to the O1 serogroup unless mentioned otherwise. Note that the transcontinental transmission event from South America to Southeast Asia (labeled “D” in reference [3], see Figure one and Supplementary Figure S3 therein) was not confirmed, and we suspect this is due to switching of the labels between strain A390 (Bangladesh 1987) and strain A316 (Argentina 1993) in that article as the positions of those strains are switched in our phylogeny.
