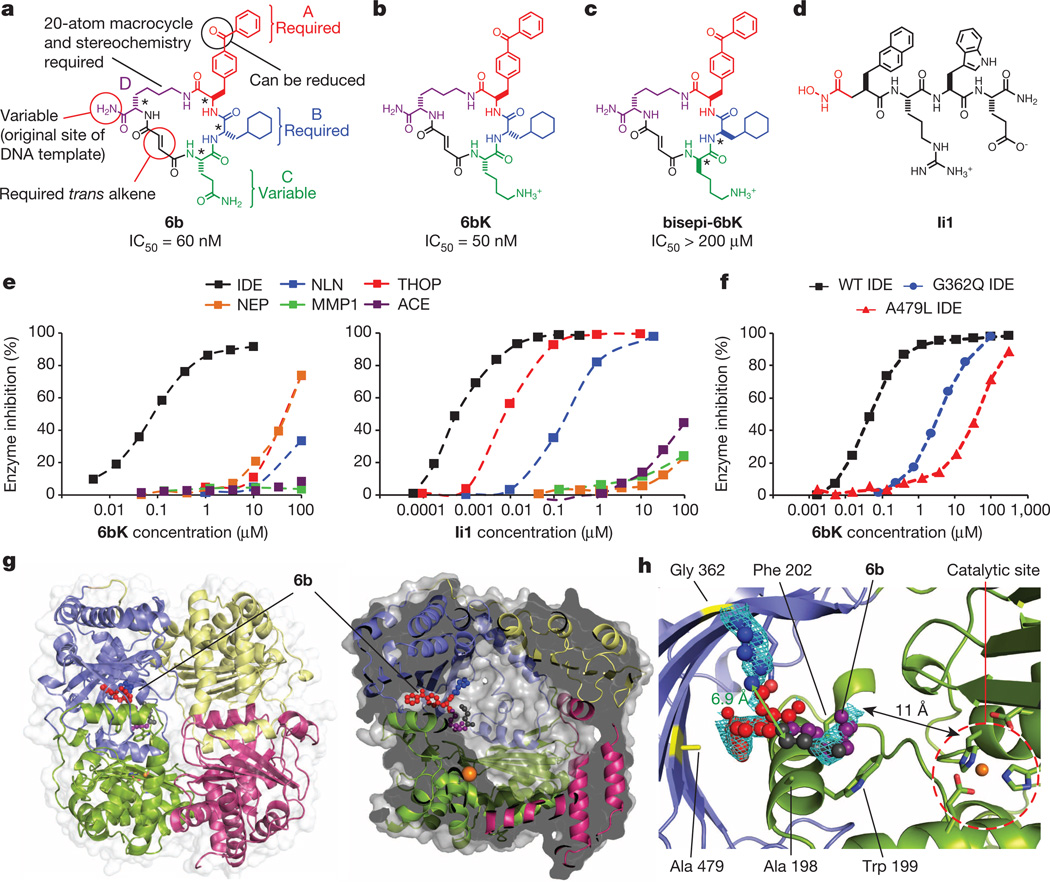
Figure 1. Potent and highly selective macrocyclic IDE inhibitors from the in vitro selection of a DNA-templated macrocycle library.
a, Structure of 6b and summary of the requirements for IDE inhibition revealed by assaying 6b analogues (Extended Data Fig. 1). b, Physiologically active IDE inhibitor 6bK. c, Inactive diastereomer bisepi-6bK. d, Previously reported substrate-mimetic hydroxamic acid Ii110. e, Selectivity analysis of macrocycle 6bK reveals >1,000-fold selectivity for IDE (IC50 = 50 nM) over all other metalloproteases tested. In contrast, inhibitor Ii110 inhibits IDE (IC50 = 0.6 nM), thimet oligopeptidase (THOP, IC50 = 6 nM) and neurolysin (NLN, IC50 = 185 nM), but not NEP (neprilysin), MMP1 (matrix metalloproteinase, 1) or ACE (angiotensin-converting enzyme). f, Activity assays for wild-type or mutant human IDE variants in the presence of 6bK. g, X-ray co-crystal structure of IDE bound to macrocyclic inhibitor 6b (2.7 Å resolution, PDB 4LTE). h, Electron density map (composite omit map contoured at 1σ) showing the relative position of macrocycle 6b bound 11 Å from the catalytic zinc atom. The glutamine residue and four atoms of the macrocycle backbone were unresolved. See also Extended Data Figs 2–4.