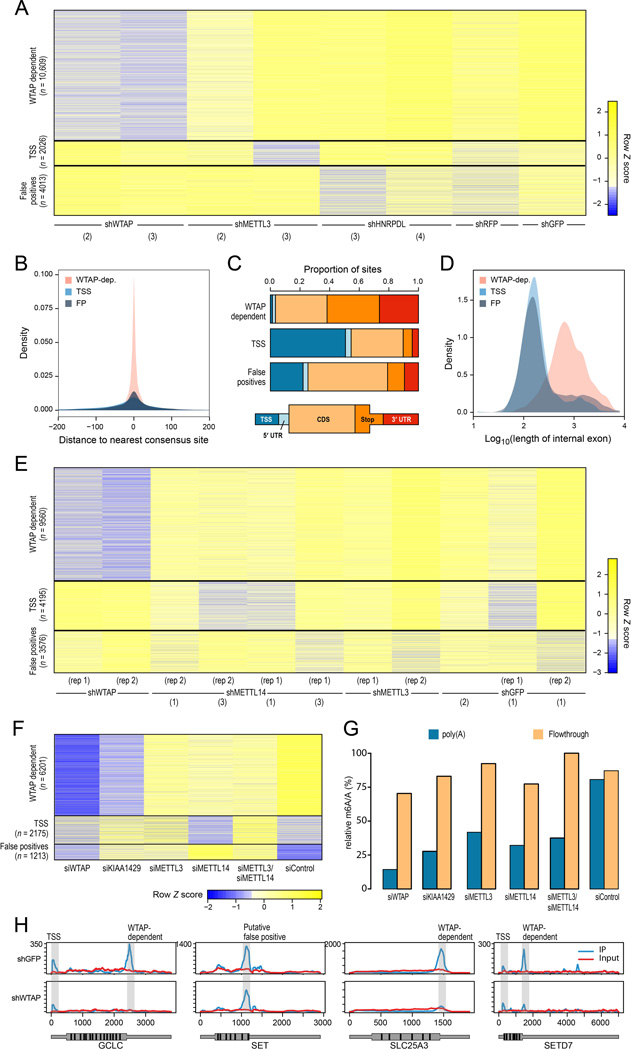
Figure 2. Perturbation of methyltransferase complex components identifies complex-dependent and -independent methylation sites.
(A) K-means clustering of log2 transformed Peak Over Input scores (yellow: high, blue: low) across 16,487 sites (rows) detected in mouse embryonic fibroblasts (MEFs). Only sites detected in 2 or more of the presented samples (columns), with a maximal Peak Over Input score greater than 8, and within genes expressed above the 60th percentile are shown. Sites were initially clustered into 5 clusters using k-means, and three of the clusters were subsequently manually merged with existing clusters, based on similarity of characteristics, to yield 3 clusters (separated by black lines). For visualization purposes, the log2(POI) scores were capped at 4.5, and then transformed into Z scores. The internal shRNA identifiers are indicated in parentheses. (B) Distributions of distance from the nearest consensus site of the sites in the three clusters in (A). (C) Distribution of the sites in (A) grouped by the clusters in (A) across five gene segments, defined as in (Dominissini et al., 2012). (D) Distribution of length of internal exons comprising the sites identified in (A) across the identified clusters. (E) K-means clustering of log2 transformed Peak Over Input scores across 17,331 sites (rows) detected in human A549 cells, visualized as in A. (F) K-means clustering of log2 transformed Peak Over Input scores for set of experiments involving perturbations of A549 cells with siRNAs, as indicated. Only WTAP-dependent sites (rows) are plotted within genes expressed above the 60th percentile across all conditions. (G) Relative m6A/A ratios across A549 cells treated with siRNA as indicated, as measured by HPLC-M/S. Values are plotted for both the poly(A) fraction (blue) and the flowthrough (orange). (H) Examples of coverage from IP (blue) and input (red) experiments across selected genes in A549 cells treated with shGFP (top row) or shWTAP (bottom row).