LIMK regulation of actin and microtubule dynamics is required for epithelial regulation of early- and late-stage cleft stabilization and progression. LIMK stimulates focal adhesion assembly and integrin β1 activation in cleft regions, causing fibronectin fibrillogenesis and promoting cleft progression during salivary gland branching morphogenesis.
Abstract
Coordinated actin microfilament and microtubule dynamics is required for salivary gland development, although the mechanisms by which they contribute to branching morphogenesis are not defined. Because LIM kinase (LIMK) regulates both actin and microtubule organization, we investigated the role of LIMK signaling in mouse embryonic submandibular salivary glands using ex vivo organ cultures. Both LIMK 1 and 2 were necessary for branching morphogenesis and functioned to promote epithelial early- and late-stage cleft progression through regulation of both microfilaments and microtubules. LIMK-dependent regulation of these cytoskeletal systems was required to control focal adhesion protein–dependent fibronectin assembly and integrin β1 activation, involving the LIMK effectors cofilin and TPPP/p25, for assembly of the actin- and tubulin-based cytoskeletal systems, respectively. We demonstrate that LIMK regulates the early stages of cleft formation—cleft initiation, stabilization, and progression—via establishment of actin stability. Further, we reveal a novel role for the microtubule assembly factor p25 in regulating stabilization and elongation of late-stage progressing clefts. This study demonstrates the existence of multiple actin- and microtubule-dependent stabilization steps that are controlled by LIMK and are required in cleft progression during branching morphogenesis.
INTRODUCTION
Branching morphogenesis is a developmental process used by several developing organs, including lungs, kidney, mammary glands, and salivary glands. The submandibular salivary gland (SMG) undergoes a series of morphodynamic transformations beginning at embryonic day 11 (E11) as the oral epithelium thickens and protrudes into the neural crest–derived mesenchyme, forming the primary bud. Multiple clefts, or tissue indentations, appear at the basal periphery of the epithelial bud and progress into the tissue interior, separating the primary bud into several smaller buds. Repeated rounds of clefting, along with tissue growth, result in a complex branched structure that undergoes further cellular and genetic changes to ultimately create a functional adult gland (Patel et al., 2006; Tucker, 2007; Knosp et al., 2012). Although many signaling pathways have been identified that regulate branching morphogenesis, the physical changes that lead to the dynamic organ-level shape changes are not clearly understood.
Cytoskeletal tension–mediated changes and spatial variations in mechanical forces set up local growth differentials that play crucial roles in tissue patterning in vivo and in vitro (Mammoto and Ingber, 2010). These mechanical cues act as intracellular signaling pathways that are required to drive changes in cell shape, cell motility, cell division, and cellular organization, permitting rapid transformations in tissue shape and integrity. Although the role of cytoskeletal elements in mechanical signal transfer has been studied in much detail in two-dimensional tissue culture systems (Parker et al., 2002; Nelson et al., 2005), their function during tissue morphogenesis remain to be fully explored. In an early study, the role of the cytoskeleton during branching morphogenesis was first identified in the mouse SMG (Spooner and Wessels, 1970, 1972). Electron micrographs of E13 explants treated with cytochalasin B showed reduced filamentous actin formation at both the apical and basal sides of the epithelial cells. Cytochalasin B treatment caused loss of clefts and inhibited morphogenesis. Of interest, when colchicine was used to inhibit microtubule polymerization, morphogenesis was halted, but the glands retained their three-dimensional (3D) shape. A model was proposed in which cleft formation was attributed to the net difference in actin-dependent contractile forces between the apical and basal cell surfaces. Microtubules were believed not to be directly required for cleft formation but were assumed to be required for general cell division (Spooner, 1973). Later studies demonstrated a role for ROCKI-stimulated nonmuscle myosin type II (myosin II)–mediated cytoskeletal contractility that was responsible for fibronectin (FN) assembly and cell proliferation driving cleft progression (Daley et al., 2009). Later it was proposed that an actin-containing ridge that regulated cleft elongation through generation of mechanical forces acted as a dynamic matrix attachment point at the base of the cleft (Kadoya and Yamashina, 2010). Although these studies implicated cytoskeletal-generated tensile forces in the process of cleft formation during branching morphogenesis, little is known regarding how the assembly/disassembly of cytoskeletal elements is coordinated during branching morphogenesis.
LIM kinases (LIMKs) control the dynamic assembly of two crucial cytoskeletal proteins, actin and microtubules (Gorovoy et al., 2005). The LIM motif–containing protein kinase family comprises two members, LIMK 1 and LIMK 2. The LIMK family contains two LIM double zinc finger domains, followed by one PDZ domain and a C-terminal catalytic domain. There is 53% overall amino acid identity between LIMK 1 and LIMK 2 (Smolich et al., 1997), and both isoforms function as dual-specificity protein kinases that phosphorylate serine/threonine and tyrosine residues. LIMK 1 and 2 have been shown to be regulated by Rho GTPases in several types of epithelial, neuronal, and tumor cells. Once activated, LIMK phosphorylates cofilin and inactivates it, preventing cofilin from severing filamentous actin and allowing accumulation of actin microfilaments. LIMK-dependent cofilin phosphorylation and subsequent dysregulation of actin affect multiple cellular functions, including actin stress fiber formation, cell migration, cell differentiation, extracellular matrix (ECM) remodeling, and metastasis (Manetti, 2012). LIMK is also known to regulate microtubule stability. Previous reports demonstrated that LIMK colocalized with tubulin in endothelial cells, and overexpression of wild-type (WT) LIMK1 induced microtubule destabilization, requiring the PDZ domain (Gorovoy et al., 2005). Another target of LIMK is tubulin polymerization–promoting protein, TPPP/p25 (Acevedo et al., 2007; Heng et al., 2012). TPPP/p25 was identified as the first member of an “intrinsically unstructured” family of unfolded proteins in oligodendrocytes in the brain, functioning as a microtubule-binding protein (Ovádi and Orosz, 2009), but it is now understood to be ubiquitously expressed (Acevedo et al., 2007). LIMK-mediated phosphorylation of p25 on a serine residue inhibits its innate ability to assemble microtubules (Acevedo et al., 2007). Thus LIMK can act as a signaling node, coordinating several cellular- and tissue-level responses during morphogenetic processes by regulating both actin filament and microtubule assembly.
In this study, we investigated a novel function for LIMK-mediated control of cytoskeletal dynamics during branching morphogenesis, using organotypic ex vivo cultures of embryonic mouse submandibular salivary glands. The function of LIMK 1 and 2 and downstream mediators cofilin and p25 were investigated using small interfering RNA (siRNA) knockdown and a small-molecule LIMK inhibitor in mouse SMG explants. Using live imaging and imaging of fixed glands subjected to immunocytochemistry and confocal imaging, we investigated a function for LIMK signaling in the formation of basement membrane and in the formation of clefts during SMG branching morphogenesis.
RESULTS
LIMK 1 and 2 are involved in the control of embryonic salivary gland cleft formation during branching morphogenesis
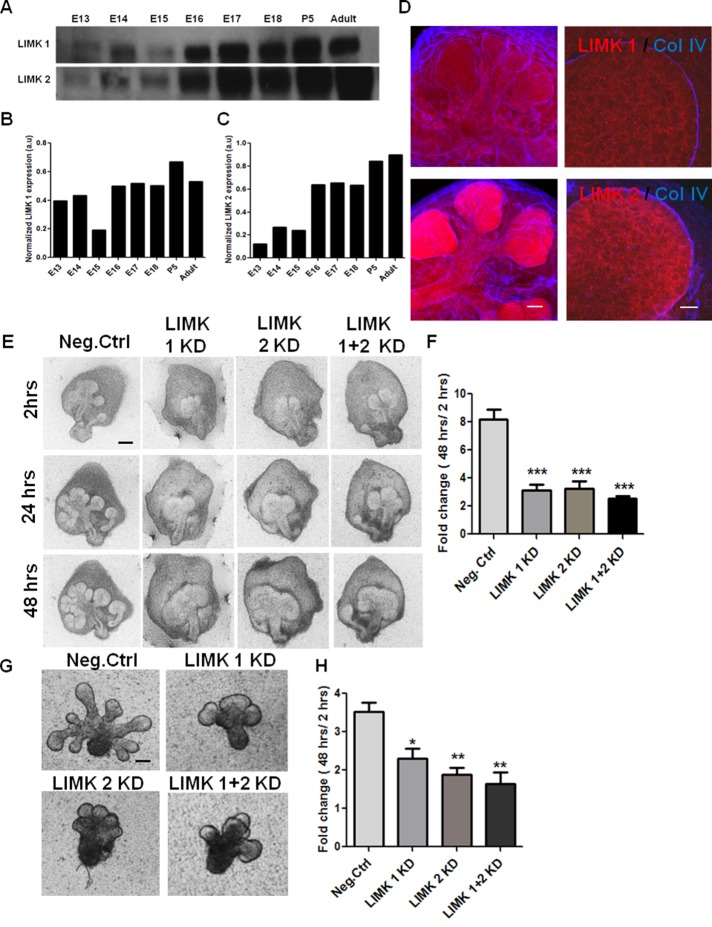
Cytoskeletal dynamics is critical for SMG branching morphogenesis (Spooner and Wessels, 1970, 1972). Because LIMK is a pivotal regulator of cytoskeletal dynamics (Manetti, 2012), we investigated the role of LIMK during SMG branching morphogenesis. We analyzed protein expression of LIMK 1 and 2 in developmental stages of mouse SMG, as shown in Figure 1A. Both LIMK 1 and 2 were highly expressed from E16 to adult stages, and moderate levels of the proteins were found in E13–E15 (Figure 1, B and C). Because the LIMK isoforms are differentially expressed in different cell types (Foletta et al., 2004; Acevedo et al., 2006) and differ in subcellular localization (Manetti, 2012), we performed immunocytochemistry to localize them in intact E13 SMGs. LIMK 1 was diffusely localized in both the epithelium and the mesenchyme, and LIMK 2 was predominantly located in the epithelium, as shown in Figure 1D. LIMK1 and 2 may thus both have roles in SMG development.
FIGURE 1:
LIMK 1 and 2 are required during early development of salivary glands. (A) Western blot analysis and densitometric quantification of total SMG protein extracts shows LIMK 1 and 2 isoforms to be present in the embryonic stages E13–E18, postnatal day 5 (P5), and adult. (B, C) LIMK 1 and 2 protein expression levels quantified relative to GAPDH levels, respectively. (D) E13 SMG organ explants were grown for 24 h and subjected to ICC/confocal imaging. LIMK 1 (red, top) is in both epithelium and mesenchyme, outside the basement membrane, as indicated by collagen IV (blue). LIMK 2 (red, bottom) is found primarily in the epithelium. Scale, 50 μm (left, 20×), 20 μm (right, 63×). (E) E13 SMGs were treated with LIMK1, 2, or 1 + 2 siRNA and subjected to bright-field imaging. Scale, 200 μm. (F) Morphometric analysis shows a significant reduction in bud number (fold change 48 h/2 h) with LIMK 1, 2, or 1 + 2 siRNA relative to negative control (n = 15). (G) Mesenchyme-free E13 epithelial rudiments treated with LIMK 1, 2, or 1 + 2 siRNA also show a decrease in branching after 48 h. Scale, 200 μm. (H) Morphometric analysis of fold changes in bud number reveals a reduction in completed buds in LIMK 1, 2, or 1 + 2 siRNA–treated glands at 48 h (n = 10). *p < 0.05, **p < 0.01, ***p < 0.001, ANOVA.
To determine whether LIMK isoforms affected cleft formation during branching morphogenesis, we treated E13 SMGs with isoform-specific LIMK siRNAs. Negative control siRNA–treated glands underwent repeated rounds of branching that yielded a highly ramified structure after 48 h, as shown in Figure 1E. Knockdown (KD) of either LIM kinase 1 or LIMK 2 (41% KD for LIMK 1 and 51% KD for LIMK 2; Supplemental Figure S1A) resulted in a morphology characterized by decreased branching relative to negative control glands. Morphometric analysis demonstrated that there are fewer buds in LIMK 1 and 2 siRNA–treated glands (Figure 1F), along with a significant reduction in the number of initiated clefts after 24 h, relative to negative control glands (Supplemental Figure S1D). Combination of equal amounts of LIMK1 and 2 siRNA produced no additional significant reduction in branching morphogenesis, suggesting that inhibition of either isoform is sufficient to affect cleft formation. To determine whether LIMK signaling is active within the epithelium, we knocked down LIMK in cultured E13 epithelial rudiments lacking mesenchyme that were embedded in a basement membrane–derived extracellular matrix (Matrigel). As with the intact SMGs, LIMK 1 and 2 KD reduced the number of completed buds in epithelial rudiments after 24 h, as observed with bright-field microscopic images and morphometric analysis (Figure 1, G and H). These findings thus establish the presence and importance of LIMK 1 and 2 in epithelial regulation of branching morphogenesis.
LIMK-mediated regulation of actin dynamics is essential for cleft formation
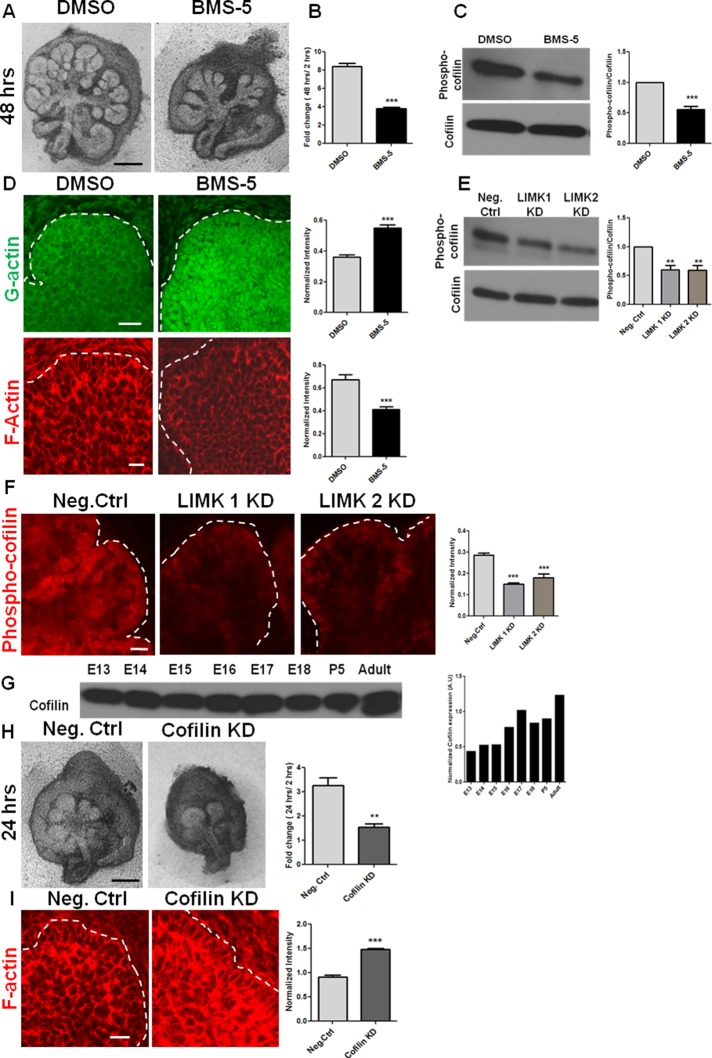
To investigate LIMK-mediated regulation of actin assembly during branching morphogenesis, we used a small-molecule inhibitor of LIMK that specifically modulates its actin-dependent downstream effects, independent of the microtubule-mediated effects. BMS-5 is a phenyl-pyrazolyl-thiazolyl-amide–substituted molecule that is known to be a highly selective and potent inhibitor of both LIMK 1 and 2 (Scott et al., 2010; He et al., 2012). When we treated E13 SMGs with 15 μM BMS-5 for 48 h, the treated glands underwent significantly fewer rounds of branching and displayed elongated buds relative to vehicle-treated glands, as demonstrated in Figure 2A. Morphometric analysis revealed a significant reduction in the number of completed buds, as seen in Figure 2B, and also in the number of stable initiated clefts after 24 h, relative to negative control glands (Supplemental Figure S1D). Because LIMK is known to inhibit the actin-severing activity of cofilin (Yang et al., 1998; Sumi et al., 1999), we examined the levels of G- and F-actin. Immunocytochemistry and confocal imaging revealed an increase in G-actin and a concomitant loss of cortical F-actin distribution in the epithelium of the BMS-5–treated explants (Figure 2D). We also examined the activation state of cofilin in the presence and absence of LIMK, using an antibody to detect cofilin that has been phosphorylated at Ser-3 and inactivated. Phosphorylated cofilin is localized intensely at the basal periphery of the epithelium and at the side walls of the cleft (Figure 2F), as detected with immunocytochemistry and confocal imaging. BMS-5 and both LIMK siRNAs stimulated a significant decrease in levels of phosphorylated cofilin, as examined through immunochemistry and Western blots (Figure 2, C, E, and F, and Supplemental Figure S2A). Because LIMK is known to affect actin stability through cofilin, we next questioned how cofilin KD affects cleft formation. Cofilin was found to be expressed abundantly throughout the E13 to adult stages, as shown in Figure 2G. When E13 SMGs were treated with cofilin siRNA (40% KD; Supplemental Figure S1B), a considerable loss in branched epithelial architecture was observed. A significant reduction in the number of completed buds was seen, as well as a reduction in stable initiated clefts, as calculated through morphometric analysis (Figure 2H and Supplemental Figure S1D, respectively). Cofilin knockdown induced an increase in F-actin, as detected with rhodamine–phalloidin in the explants (Figure 2I). These results demonstrate that alteration in actin stability, by either increasing (cofilin KD) or decreasing (LIMK inhibition by BMS-5) the proportion of filamentous actin in the salivary epithelium, causes a significant perturbation in branching morphogenesis.
FIGURE 2:
Actin dynamics regulated by LIMK and cofilin is required for cleft formation. (A) E13 SMG organ explants were grown for 48 h ± a pharmacological inhibitor of LIMK, BMS-5. Scale, 200 μm. (B) Morphometric analysis shows a significant reduction in the fold change in buds in the presence of BMS-5 after 48 h of incubation; n = 15, paired t test. (C) Quantification of Western blots of E13 SMGs extracts treated with BMS-5 for 48 h confirm LIMK-mediated inhibition of cofilin phosphorylation at Ser-3; n = 4, paired t test. (D) Confocal images of ICC for G-actin (green) and F-actin (red) after 48 h of BMS-5 treatment in E13 SMGs. Quantification of multiple images shows increased G- and reduced F-actin in the epithelium of BMS-5–treated glands. Scale, 20 μm; n = 5, paired t test. (E) Quantification of Western blots of E13 SMG treated with LIMK 1 or 2 siRNA and examined for cofilin phosphorylation at Ser-3 shows reduced levels of phosphorylated cofilin with LIMK KD; n = 4, ANOVA. (F) Immunocytochemistry and confocal imaging detects phospho-ser3-cofilin localized in the cleft region of untreated E13 glands and decreased phospho-ser3-cofilin in LIMK 1 and 2 siRNA–treated E13 glands after 48 h. Quantification of images from multiple SMGs indicates overall reduction of phospho-cofilin levels with LIMK KD. Scale, 20 μm; n = 5, ANOVA. (G) Western blot and densitometric quantification show cofilin to be present in the developmental stages E13–E18, P5, and adult. (H) E13 SMG organ explants were grown for 24 h ± cofilin siRNA. Morphometric analysis shows a reduction in completed buds after 24 h. Scale, 200 μm; n = 6, paired t test. (I) Confocal images of ICC for F-actin (red) of E13 SMGs treated for 24 h ± cofilin siRNA. Quantification of images from multiple SMGs shows increased F-actin with cofilin KD. Scale, 200 μm; n = 4, paired t test. **p < 0.01, ***p < 0.001.
A consequence of altered actin assembly is changes in cell shape. LIMK has been found to affect tumor cell morphology in 3D matrices (Mishima et al., 2010). We therefore questioned whether LIMK inhibition had any significant effect on cell shape during organogenesis. The embryonic epithelial bud at E13 is organized into outer cuboidal cells (OCCs) near the basal periphery and inner polymorphic cells (IPCs) in the interior of the bud (Daley et al., 2012). Under LIMK inhibition with BMS-5, the organizational distinction between OCCs and IPCs was lost (Supplemental Figure S2B) with a decrease in both the cell perimeter (Supplemental Figure S2C) and area (Supplemental Figure S2D). To confirm that BMS-5 was not toxic, we performed inhibitor washout experiments after 24 h of BMS-5 treatment. E13 SMGs showed significant structural recovery through increased branching (Supplemental Figure S3A), formation of acinar units with cortical localization of F-actin (Supplemental Figure S3D), and increased levels of phospho-cofilin (Ser-3; Supplemental Figure S3, B and C) after 48 h of washout. BMS-5 did not induce cell death, as observed by cleaved caspase-3 staining (Supplemental Figure S2F). Taken together, these results demonstrate that LIMK is able to significantly alter cell shape through reorganization of actin filaments during branching morphogenesis in the mouse salivary gland.
Regulation of microtubule dynamics by TPPP/p25 and LIMK is required during branching morphogenesis
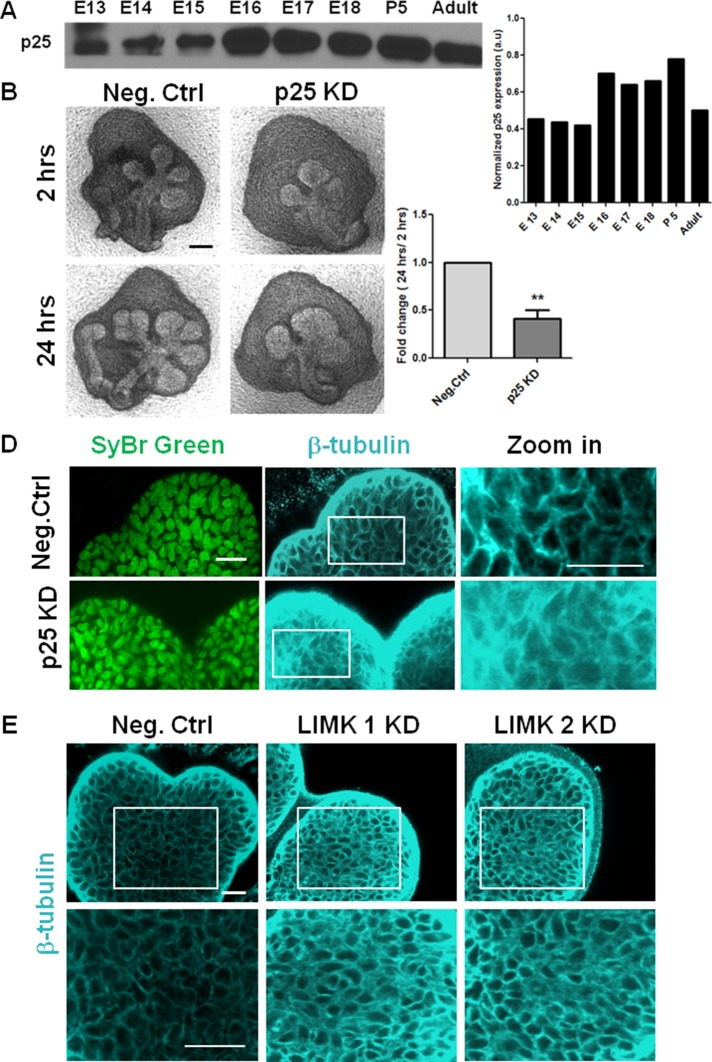
Because LIMK also regulates microtubule stability, we hypothesized that LIMK could contribute to morphogenesis through regulation of microtubule organization. LIMK 1 phosphorylates p25 on a serine residue in vitro, and this inactivation of p25 leads to microtubule disassembly in vitro and in NIH3T3 cells (Acevedo et al., 2007). LIMK 2 also interacts with p25 during astral microtubule organization in HeLa cells (Heng et al., 2012). We examined p25 protein levels in mouse salivary glands at multiple developmental time points and found that it is present at moderate amounts in E13–E15 (Figure 3A). Knocking down p25 with a specific siRNA (54% knockdown at the protein level; Supplemental Figure S1C) led to a significant reduction in branching morphogenesis (Figure 3B), and morphometric analysis revealed a substantial lack of completed buds (Figure 3C). Because p25 is known to control the distribution and localization of β-tubulin, we examined its effect on microtubules using ICC and confocal imaging in p25 siRNA–treated E13 SMGs. p25 KD induced a diffuse distribution of β-tubulin in the epithelium that obscured the distinct cell borders apparent in the negative control–treated glands (Figure 3D). In comparison, glands treated with LIMK 1 and 2 KD exhibited thick filamentous microtubules around the periphery of the cells throughout the epithelium as compared with negative control siRNA–treated glands (Figure 3E). When SMGs were treated with Taxol, a similar increase in β-tubulin staining intensity, as seen with LIMK 1 and 2 KD, was observed as a result of overstabilization of microtubules (Supplemental Figure S2E). These data demonstrate that LIMK-mediated microtubule stability is essential during branching morphogenesis and suggest that this function may be mediated through p25. Formation of new buds thus requires a fine organizational balance of microtubules and aberrant disassembly (via p25 KD) or assembly (via LIMK KD) of microtubule network is detrimental to branching morphogenesis in the mouse salivary gland.
FIGURE 3:
Regulation of microtubule dynamics by TPPP/p25 and LIMK is required during branching morphogenesis. (A) Western blot analysis and densitometric quantification show developmental expression of p25 protein from E13 to E18, P5, and adult. (B) E13 SMG organ explants were grown for 24 h ± p25 siRNA and imaged with bright-field microscopy. Scale, 200 μm. (C) Morphometric analysis shows a significant reduction in buds with p25 KD; n = 10, paired t test, **p < 0.01. (D) Mesenchyme-free E13 SMG rudiments were grown for 24 h ± p25 siRNA, stained for SyBr green (green), and subjected to ICC for β-tubulin (cyan) and imaged with confocal microscopy. In p25 KD, β-tubulin exhibits a diffuse distribution in the epithelium relative to negative control siRNA–treated glands (as seen in zoomed-in image), indicative of reduced microtubule stability. Scale, 20 μm. (E) Mesenchyme-free E13 SMG rudiments were grown for 48 h ± LIMK 1 or 2 siRNA vs. negative control siRNA. LIMK 1 and 2 KD demonstrated increased β-tubulin staining at the cell borders (bottom, zoomed-in area) in the epithelium relative to negative control siRNA–treated glands, indicative of overstabilized microtubules. Scale, 20 μm.
LIMK inhibition exerts multistep control on cleft formation via disruption of actin and microtubule dynamics
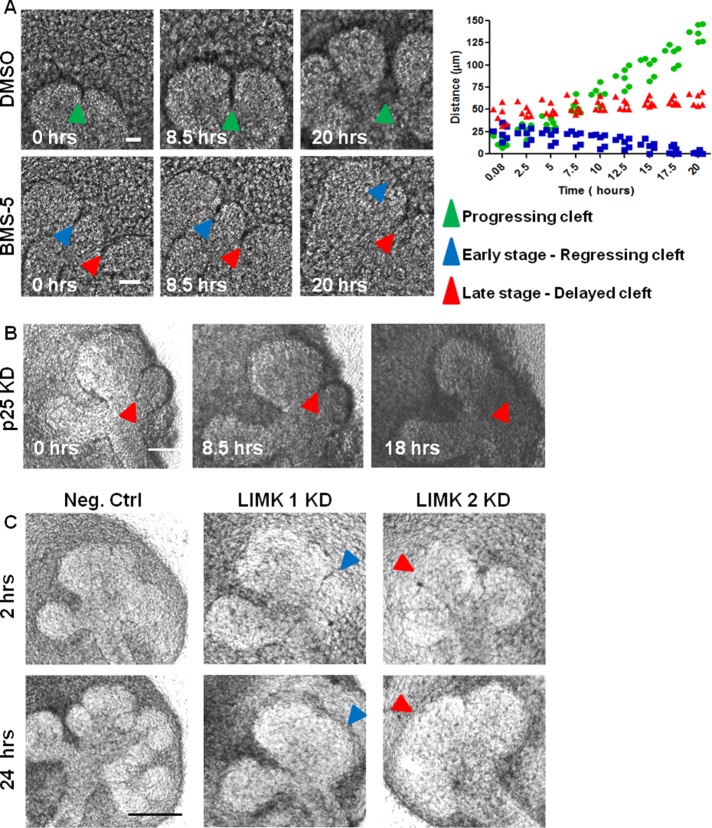
To specifically examine the role of cytoskeletal stability on cleft progression, we performed time-lapse microscopy on BMS-5–, LIMK siRNA–, cofilin siRNA–, and p25 siRNA–treated glands. In vehicle control–treated epithelium (dimethyl sulfoxide [DMSO]), initiated clefts continue to progress over time and terminate with a widened base, as exemplified in Figure 4A (progressing cleft; green arrow). In contrast, in BMS-5–treated epithelium, initiated clefts that were <40–45 μm in length were gradually lost during 20 h of incubation (early stage, regressing cleft; blue arrow). This cleft regression is consistent with the previous observation that cytochalasin B causes loss of clefts, although the length of the lost clefts was not reported (Spooner and Wessels, 1972). In this study, clefts that were >45 μm in length persisted but progressed very slowly compared with clefts in vehicle-treated controls (late stage or delayed cleft; red arrow). When cleft progression was plotted against time, the rate of cleft growth in vehicle-treated glands was 6.8 μm/h, but for BMS-5–treated glands (late stage or delayed cleft), it was only 3.1 μm/h. Time-lapse microscopy with cofilin KD also showed that short progressive clefts elongate very slowly and display lack of termination under 20 h of incubation with siRNA, as seen in Supplemental Figure S1E. Time-lapse microscopy with p25 KD revealed that progression-competent clefts as long as 65 μm regressed in 24 h. Only those clefts that were near termination and starting to widen at the base were retained, as shown in Figure 4B. With LIMK 1/2 siRNA, which regulates both actin and microtubule stability, not only smaller clefts, but also clefts as long as 65–70 μm, were found to regress in 24 h. However, longer clefts that had widened, yielding subdivision of the epithelial bud, were retained, as shown in Figure 4C. Taken together, these data establish a requirement for actin in the early stages of cleft stabilization and progression, whereas dynamic regulation of microtubules appears to be required mainly at the later stages of cleft formation.
FIGURE 4:
LIMK knockdown and inhibition result in lack of cleft stabilization and inhibition of cleft progression. (A) E13 SMGs were treated ± BMS-5 and subjected to bright-field time-lapse imaging. Images captured at times indicated a complete cycle of cleft initiation, progression, and termination (green arrowhead) in vehicle-treated (DMSO) SMGs. With BMS-5 treatment, early-stage clefts regress (blue arrowhead) and later-stage clefts show slower progression (red arrowhead). Distance vs. time plot for clefts in vehicle control– and BMS-5–treated glands show different patterns for cleft progression for multiple clefts (n = 12). Scale, 50 μm. (B) E13 SMGs treated with p25 siRNA and subjected to bright-field time-lapse imaging demonstrate that p25 KD causes regression of late-stage progressive clefts (blue arrowheads). Scale, 100 μm. (C) E13 SMGs were treated with LIMK 1 or 2 siRNAs and subjected to bright-field time-lapse imaging. LIMK 1 and 2 KD reveals regression of both early-stage (blue arrowheads) and late-stage (red arrowheads) clefts after 24 h of incubation. Scale, 200 μm.
LIMK regulation of cell proliferation is required for branching morphogenesis
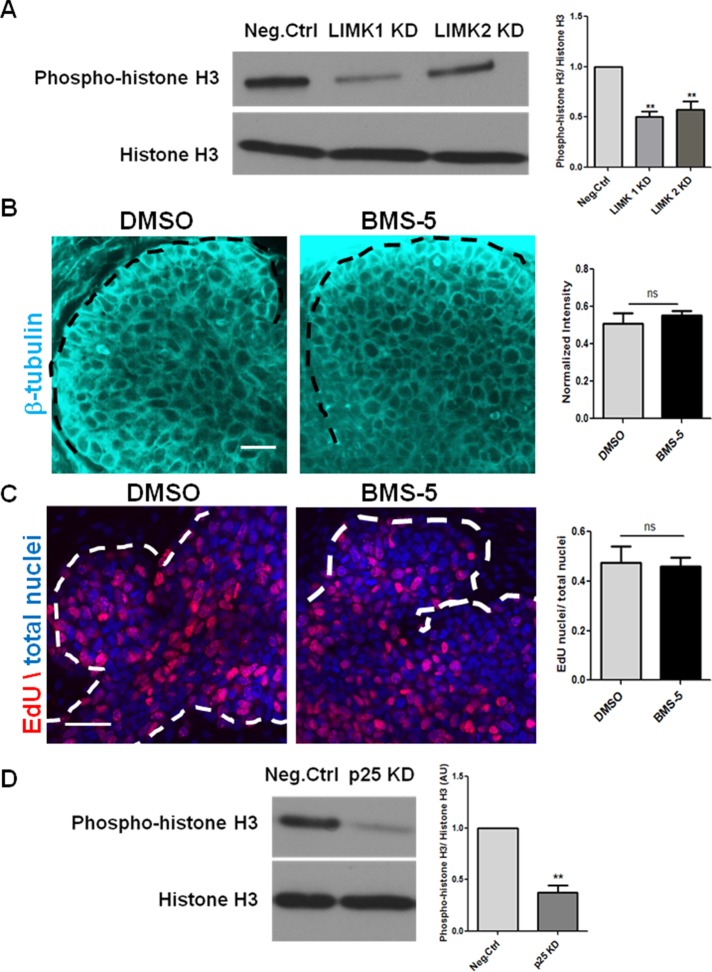
Because cell proliferation is a major contributor to branching morphogenesis, we investigated the change in cell proliferation in LIMK-inhibited glands. Although cell proliferation is required for branching, clefts can still form when proliferation is inhibited (Nakanishi et al., 1987). Cleft progression does require cell proliferation, which is, in part, stimulated by FN (Daley et al., 2009). When LIMK-inhibited E13 glands were examined for proliferation, it was apparent that LIMK KD significantly reduced phosphorylation of histone H3 (pHH3), an M-phase marker, relative to negative control SMGs (Figure 5A). As reported previously (Ross-Macdonald et al., 2008), BMS-5 did not affect the organization or stability of microtubules, as detected with β-tubulin immunocytochemistry (Figure 5B), and also did not affect cell proliferation, as detected with 5-ethynyl-2′-deoxyuridine (EdU)–positive cells in S phase (Figure 5C). With p25 KD, which caused microtubules to disassemble in E13 SMGs, it was observed that there was no overall growth in the tissues. Western blot analysis detected a significant decrease in cell proliferation with reduced pHH3 levels in glands treated with p25 siRNA (Figure 5D). These data are consistent with the hypothesis that proper control of microtubule organization by LIMK and p25 is required for regulation of proliferation in the developing SMG.
FIGURE 5:
LIMK regulates epithelial proliferation during branching morphogenesis. (A) E13 SMGs were treated with LIMK 1 or 2 siRNA for 48 h and subjected to Western analysis. Densitometric quantification reveals that LIMK 1 and 2 KD reduces proliferation, as detected by phospho–histone 3 (pHH3), relative to negative control siRNA–treated glands; n = 3, **p < 0.01, ANOVA. (B) E13 SMGs were treated with BMS-5 for 48 h and subjected to ICC/confocal imaging to detect β-tubulin. Scale, 20 μm. Quantification reveals that BMS-5 did not alter staining intensity or epithelial localization of β-tubulin relative to DMSO-treated glands; n = 4, paired t test. (C) Quantification of fluorescence intensity reveals that BMS-5 did not alter the proliferative capacity of epithelial cells relative to vehicle control–treated glands, as detected with EdU staining (red) relative to total nuclei (SyBr Green, pseudo colored blue). Scale, 20 μm; n = 3, paired t test. (D) Western blot and densitometric analysis shows p25 siRNA reduces pHH3 in comparison to negative control siRNA–treated glands after 24 h; n = 4, **p < 0.01, paired t test.
LIMK regulates cleft stabilization through focal adhesion activation and hence FN assembly in an integrin-dependent manner
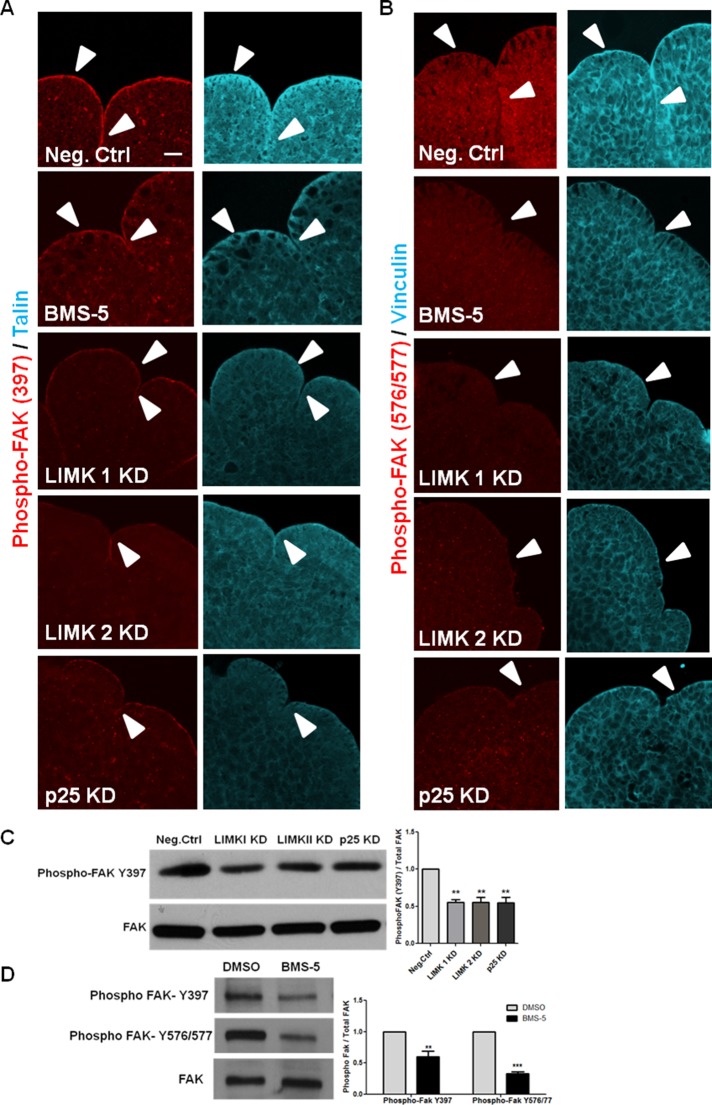
Focal adhesions (FAs) are multimeric protein complexes that function as mechanosensors through integrins to mediate bidirectional interactions between the cytoskeleton and ECM proteins (Wozniak et al., 2004). Of interest, LIMK1 was previously found to affect focal adhesion activation in fibroblasts and breast cancer cells (Foletta et al., 2004; McConnell et al., 2011), and overexpressing WT LIMK 1 and 2 caused an increase in the size and number of focal adhesion complexes (Sumi et al., 1999). Because both LIMK siRNA- and BMS-5–treated glands showed regression of progressive clefts along with a lack of stable initiated clefts 24 h after culture, we hypothesized that LIMK-mediated stimulation of focal adhesion assembly is responsible for regulating cleft stability during cleft progression. In E13 epithelial rudiments, phosphorylated FAK and the adaptor proteins localized primarily at the basal periphery of the epithelium and along the sides of the cleft walls under control conditions (Figure 6, A and B). When E13 epithelial rudiments were treated with BMS-5, LIMK 1, LIMK 2, or p25 siRNA, we found a decrease in levels of phosphorylated FAK at both the autophosphorylated and Src-phosphorylated tyrosines (Y397 and Y576/577, respectively) in the cleft region, along with a lack of recruitment of the adaptor proteins talin and vinculin, relative to negative control siRNA–treated glands. Cofilin KD was also found to reduce the levels of phosphorylated FAK (Y397 and Y576/577), vinculin, and talin in the cleft regions (Supplemental Figure S3, E and F). Western analysis in intact E13 SMGs treated with LIMK siRNA showed a 44% reduction, and SMGs treated with p25 siRNA showed a 47% reduction in phosphorylated FAK (Y397; Figure 6C). With BMS-5 treatment, a 46% reduction in phosphorylated FAK (Y397) and a 67% decrease in phosphorylated FAK (Y576/77) relative to DMSO-treated glands was observed (Figure 6D), consistent with the ICC data. A minor reduction in phosphopaxillin at Tyr-118 was observed in LIMK and p25 KD glands (Supplemental Figure S2I), with almost no reduction in the presence of BMS-5 (Supplemental Figure S2, G and H). Together these data indicate that LIMK-mediated assembly of focal adhesion protein–containing complexes depend on dynamic assembly/disassembly of both actin filaments and microtubules.
FIGURE 6:
LIMK-mediated cytoskeletal organization is necessary for focal adhesion protein localization in the cleft region of SMGs. (A) E13 epithelial rudiments were cultured for 24 h with negative control siRNA, BMS-5, LIMK 1, 2, or p25 siRNAs. Glands were immunostained for FAK phosphorylated on Tyr-397 (red) and the focal adhesion protein talin (cyan) or (B) phosphorylated FAK (Tyr-576/577) (red) and vinculin (cyan). In negative control rudiments, pFAK Y397 and pFAK (Y576/577) with the focal adhesion protein talin and vinculin localized to the periphery of epithelial buds (white arrow head) and along the side walls of progressing clefts. Both pFAK Y397 and Y576/577, as well as talin and vinculin levels, were reduced with LIMK 1/2 and p25 siRNA and BMS-5. Scale, 20 μm. (C, D) E13 SMG treated with LIMK 1, 2 and p25 siRNA and BMS-5 were immunoblotted with antibodies specific for pFAK Tyr-397 or Tyr-576/577. A decrease in phosphorylation at both sites was detected after quantification relative to total FAK protein; n = 3, **p < 0.01, ***p < 0.001, ANOVA.
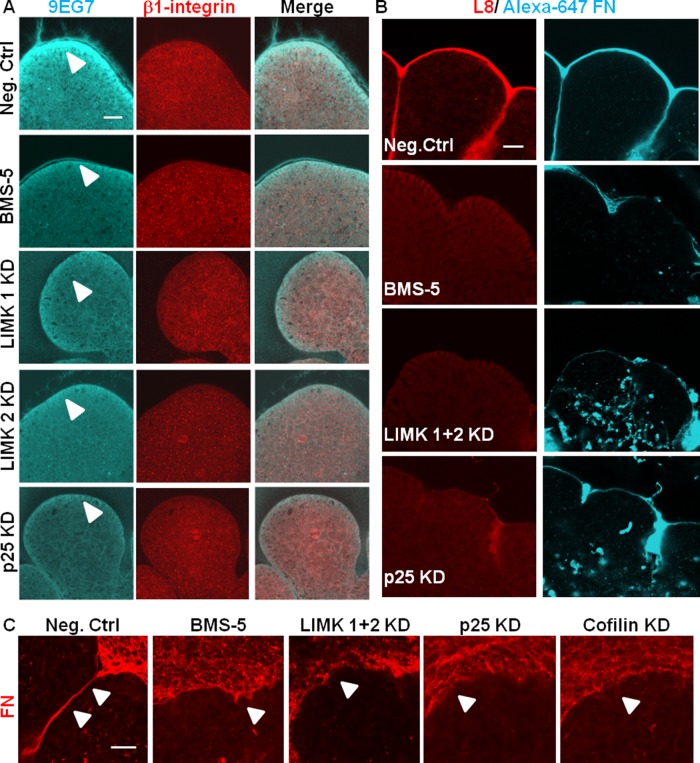
Because integrin β1 is required for branching morphogenesis (Sakai et al., 2003), we assayed for β1-integrin activation in the LIMK-inhibited SMG explants. Mesenchyme-free E13 epithelial rudiments were treated with the LIMK inhibitor BMS-5 and LIMK and p25 siRNAs. Integrin activation was assayed using the 9EG7 monoclonal antibody to detect active integrin β1 relative to total integrin β1. 9EG7 was found to stain most intensely at the basal periphery of the epithelial buds (Figure 7A), whereas β1 integrin antibody stained diffusely in the entire epithelium. With LIMK inhibition and knockdown, although there was no change in total integrin β1 staining intensity, there was a significant decrease in integrin β1 activation compared with control SMGs. As shown in Figure 7A, 9EG7 staining decreased in the basolaterally located epithelial cells in BMS-5–treated SMGS and also in LIMK 1, LIMK 2, and p25 siRNA–treated SMGs. These data suggest that the reduced activation of integrin β1 with inhibition of LIMK can be attributed to the perturbation of the dynamic turnover of microtubules and actin.
FIGURE 7:
LIMK regulates integrin activation and FN assembly. (A) Mesenchyme-free E13 epithelial rudiments were cultured in the presence of negative control, BMS-5, LIMK 1, LIMK 2 siRNA, or p25 siRNA, costained with a polyclonal antibody recognizing total integrin β1 (red) and active integrin β1 (mAb 9EG7), and examined with confocal imaging. Active integrin β1 (cyan) is detected primarily at the basal periphery of the epithelial buds (white arrowhead), and the staining intensity is reduced in LIMK inhibitor– and siRNA–treated glands, whereas total integrin β1 is unchanged. Scale, 20 μm. (B) Mesenchyme-free E13 epithelial rudiments were cultured in the presence of Alexa 647–labeled human plasma FN (cyan) and treated with negative control siRNA, BMS-5, LIMK 1 + 2 siRNA, or p25 siRNA and subjected to ICC to detect assembled fibrillar FN (L8 antibody, red) by confocal imaging. BMS-5–, LIMK 1 + 2 siRNA–, and p25 siRNA–treated SMGs show a reduction in assembled FN (red) around the epithelial periphery relative to negative control–treated SMGs. Aberrant accumulation of FN (cyan) within the epithelium is observed in LIMK 1 + 2 and p25 siRNA–treated glands. Scale, 20 μm. (C) Intact E13 SMGs were cultured in the presence of negative control siRNA, BMS-5, LIMK 1 and 2 siRNA, p25 siRNA, or cofilin siRNA and subjected to ICC/confocal imaging to detect FN. BMS-5–, LIMK 1 and 2 siRNA–, p25 siRNA–, and cofilin siRNA–treated SMGs show disorganized and uneven deposition of FN (red) in the basement membrane surrounding the epithelium. Scale, 20 μm.
FN has been long known to regulate cleft progression, during which epithelial cell–cell adhesions are continuously replaced by cell–matrix adhesions as FN is assembled in the progressing clefts (Sakai et al., 2003; Larsen et al., 2006; Onodera et al., 2010). Because BMS-5–treated clefts displayed significantly reduced rates of progression, we hypothesized that LIMK regulation of FN matrix assembly is required for efficient cleft progression. LIMK 1 was previously shown to promote FN-mediated cell attachment through focal adhesion formation in two-dimensional cell culture (Horita et al., 2008). Under conditions of LIMK inhibition, LIMK KD, cofilin KD, and p25 KD, intact organ explants were immunostained with an anti-FN antibody, which showed disorganized and uneven FN staining in the basement membrane region (Figure 7C). We used mesenchyme-free epithelial rudiments and pretreated them with a low amount of Alexa 647–labeled purified human plasma FN sufficient to label the basement membrane without significantly affecting branching morphogenesis. When E13 epithelial rudiments pretreated with labeled plasma FN were incubated with BMS-5, LIMK 1 + 2 siRNA, or p25 siRNA, significant differences in FN matrix organization were observed in comparison with control SMGs. In negative control siRNA–treated SMGs, substantial deposition of FN matrix, as labeled by Alexa 647–FN (Figure 7B), was detected. In contrast, significantly decreased FN deposition in the basement membrane was detected in BMS-5–treated glands. In addition, ectopic deposition of FN in the interior of the epithelial bud was observed in LIMK and p25 siRNA–treated glands, along with significantly reduced basal deposition at the epithelial periphery. This FN localized within the epithelial bud appeared to be predominantly extracellular, both in the mesenchyme-free rudiments and in intact SMGs treated with LIMK and p25 siRNA (Supplemental Figure S4, A and B).
To determine whether there is a defect in FN assembly in the presence of LIMK inhibition, the L8 monoclonal antibody was used, which is known to specifically bind to the cryptic site that is unmasked when integrin-bound FN undergoes conformational changes in response to cytoskeletal stress (Zhong et al., 1998). In control SMGs, L8 immunoreactivity was detected primarily within the basement membranes of epithelial tissues, colocalizing with Alexa 647–FN. In LIMK and p25 knockdown glands, such reactivity was markedly reduced, although in BMS-5–treated glands, the L8 reduction was less significant than in LIMK siRNA–treated SMGs (Figure 7B). These data are consistent with a requirement for LIMK in “inside-out” integrin activation, enabling FN assembly during cleft progression in developing SMGs through regulation of both microtubule and actin stability.
FN promotes branching in an LIMK-dependent pathway
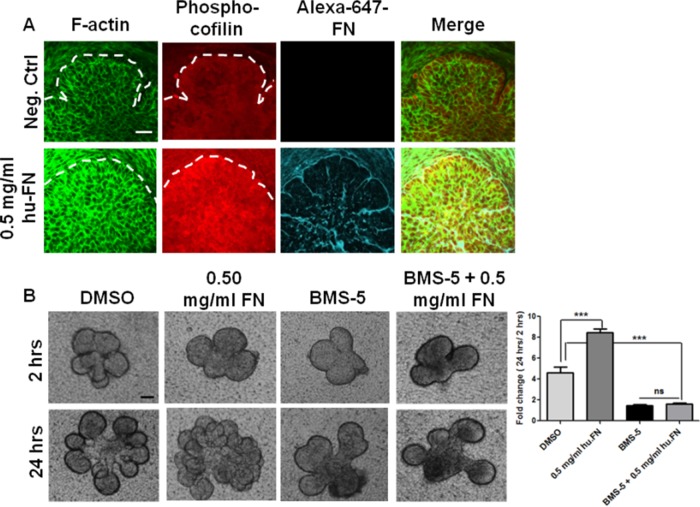
FN has been shown to increase branching morphogenesis in a dose-dependent manner (Sakai et al., 2003). This increase in branching was associated with an increase in phosphorylated FAK (Y397) levels as compared with control SMG (Supplemental Figure S3G). This increase in FAK activation correlated with an increase in actin stability, as seen with increased F-actin and phospho-ser3-cofilin levels (Figure 8A and Supplemental Figure S3G). This is consistent with the previously demonstrated effect of exogenously added FN stimulating increased branching in E13 SMGs by promoting “outside-in” signaling. Of interest, when FN was added in presence of the LIMK inhibitor BMS-5, no significant increase in the branching pattern was observed compared with BMS-5–treated and DMSO-treated epithelial rudiments (Figure 8B). These data thus show that FN must be actively assembled via a LIMK-dependent pathway to stimulate cleft formation.
FIGURE 8:
FN promotes branching in a LIMK-dependent manner. (A) Treatment of E13 SMG with 0.5 mg/ml exogenous plasma FN (cyan) for 24 h increases F-actin (green) along with an increase in cofilin phosphorylation at Ser-3 (red), as revealed by ICC/confocal imaging. Scale, 20 μm. (B) FN-stimulated branching in E13 epithelial rudiments after 24 h is prevented in the presence of BMS-5, as indicated by bright-field images and morphometric analysis of bud numbers in multiple glands. Scale, 200 μm; n = 4, ***p < 0.001, ANOVA.
Effects of LIMK knockdown are rescued by restoring actin and microtubule dynamics
Bidirectional signaling between the actin cytoskeleton and integrins (Schoenwaelder and Burridge, 1999; Calderwood et al., 2000) was previously investigated. It was observed that ECM binding to activated integrins is not sufficient for matrix assembly without the dynamic remodeling of the actin cytoskeleton (Wu et al., 1995). Because exogenous addition of FN in LIMK inhibited glands could not rescue branching morphogenesis, we hypothesized that restoration of the actin and microtubule dynamics in the presence of LIMK down-regulation is required to rescue cleft formation.
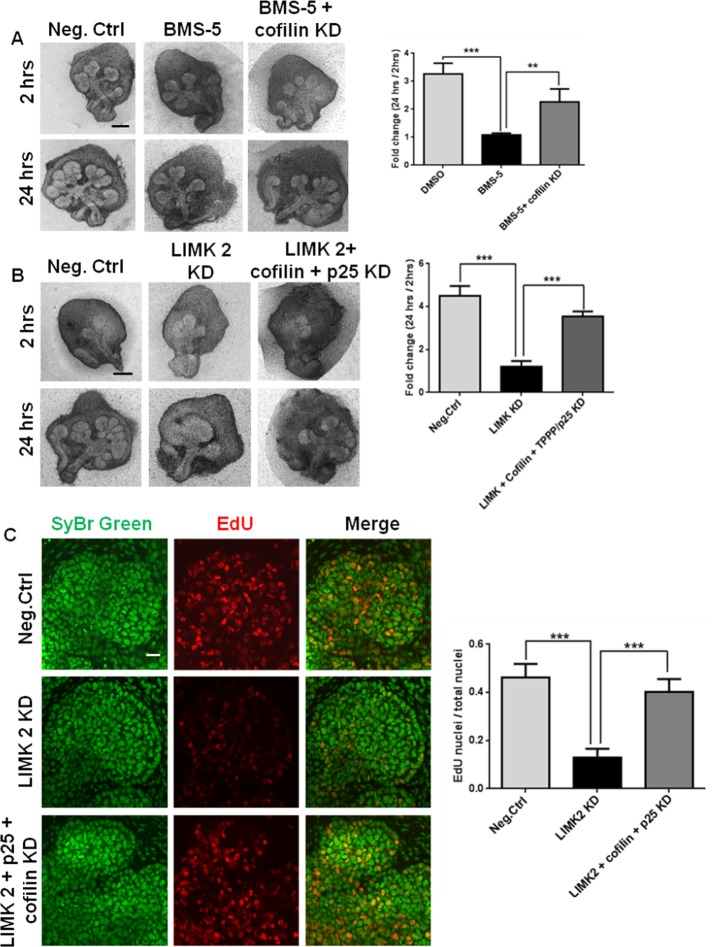
Because LIMK inhibition activates both cofilin and p25, we reasoned that knockdown of cofilin or p25 in the presence of LIMK inhibition would rescue branching morphogenesis. We therefore used cofilin siRNA to reduce cofilin levels together with the LIMK inhibitor BMS-5 for 24 h and assayed for branching morphogenesis. As shown in Figure 9A, a significant improvement in the branched morphology of the gland was observed upon addition of cofilin siRNA (34% KD; Supplemental Figure S5A) in the presence of BMS-5 as compared with BMS-5–treated glands. Because the knockdown of proteins with siRNA in organ explants does not reach 100% efficiency, a complete rescue of branching was not observed in glands treated with cofilin siRNA in the presence of BMS-5. We demonstrated that LIMK knockdown affects both actin and microtubule stability, so we attempted to rescue effects of LIMK knockdown on cleft formation with p25 siRNA (37% KD; Supplemental Figure S5B) together with cofilin siRNA, added in the presence of LIMK 2 siRNA (Figure 9B). With this triple KD, we detected an increased number of completed buds compared with LIMK2 siRNA treatment, as quantified with morphometric analysis (Figure 9B).
FIGURE 9:
LIMK-mediated inhibition of branching morphogenesis is rescued by knockdown of cofilin and p25 with up-regulation of proliferation. (A) E13 SMGs were subjected to BMS-5 ± cofilin siRNA for 24 h, imaged with bright-field imaging, and subjected to morphometric quantification of fold change in completed buds. Scale, 200 μm. LIMK inhibition of branching morphogenesis by BMS-5 is partially recovered in the presence of cofilin siRNA. The fold change in number of buds grown for 24 h shows a significant increase compared with BMS-5 treatment; n = 3, **p < 0.01, ***p < 0.001, ANOVA. (B) E13 SMGs were subjected to LIMK 2 siRNA ± cofilin and p25 siRNA for 24 h, imaged with bright-field imaging, and subjected to morphometric quantification of fold change in completed buds. Scale, 200 μm. Glands subjected to LIMK 2 KD recover morphologically in the presence of both cofilin and p25 siRNAs. The fold change in number of buds shows a significant increase in branching compared with LIMK 2 siRNA; n = 3, ***p < 0.001, ANOVA. (C) E13 SMGs were subjected to LIMK 2 siRNA ± cofilin and p25 siRNAs for 24 h, treated with EdU, and subjected to confocal imaging to detect EdU (red) vs. SyBr green (green). Scale, 20 μm. Fluorescence intensity quantification reveals that LIMK2-mediated inhibition of proliferation is rescued with cofilin and p25 siRNAs by 70%; n = 3, ***p < 0.001, ANOVA.
To confirm that the cellular and molecular responses downstream of LIMK could be restored with knockdown of cofilin and/or p25 in the presence of LIMK inhibition, we performed immunostaining for the LIMK-regulated protein targets. When p25 and cofilin siRNA were added to cultured explants in the presence of LIMK2 siRNA, the epithelial distribution of β-tubulin was found to be similar to the negative control siRNA–treated glands rather than the LIMK2 siRNA–treated glands (Supplemental Figure S6A). Because we previously demonstrated that cell proliferation is inhibited by LIMK KD through microtubule organization, we investigated the effects of cofilin and p25 KD on cell proliferation in the presence of LIMK KD. Figure 9C shows that recovery of LIMK knockdown with simultaneous addition of cofilin and p25 siRNAs in presence of LIMK KD increased the number of EdU-positive cells compared with LIMK siRNA treatment.
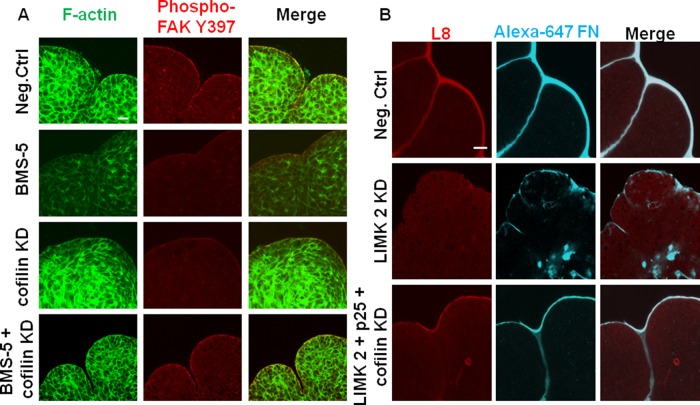
We previously demonstrated that BMS-5 and cofilin knockdown affected the actin cytoskeleton and focal adhesion assembly in opposite ways: LIMK inhibition and KD via BMS-5 or siRNA lead to decreased levels of actin polymerization, whereas cofilin KD increased levels of F-actin. In addition, phosphorylation of FAK adjacent to the basement membrane was decreased when LIMK was inhibited. We reasoned that the state of the actin cytoskeleton and localization of active FAK could be rescued by knockdown of cofilin in the presence of LIMK inhibition. When cofilin siRNA was added in the presence of BMS-5, F-actin dynamics were found to stabilize, resembling the negative control–treated explants. This restored focal adhesion kinase localization at the cleft region and phosphorylation at the Tyr397 residue (Figure 10A). A similar rescue of focal adhesion–based cleft stability with LIMK knockdown was achieved by including p25 with cofilin in presence of LIMK siRNAs, with a resulting increase in phospho-FAK (Y397) levels (Supplemental Figure S5C).
FIGURE 10:
Rescue of LIMK inhibition restores F-actin dynamics along with focal adhesion kinase activity and FN deposition and assembly in the cleft regions of embryonic glands. (A) Mesenchyme-free E13 epithelial rudiments treated with BMS-5, cofilin siRNA, or BMS-5 plus cofilin siRNA for 24 h were subjected to ICC/confocal imaging to detect pFAK (Y397, red) vs. F-actin (green). The decreased F-actin with LIMK inhibition by BMS-5 and increased F-actin with cofilin siRNA both disrupted FAK activation in the cleft region. Cofilin KD in the presence of BMS-5 stabilized actin dynamics and restored downstream FAK activation. Scale, 20 μm. (B) Mesenchyme-free E13 epithelial rudiments treated with control siRNA or LIMK2 ± cofilin and p25 siRNAs for 24 h were grown in the presence of Alexa 647–labeled human plasma FN (cyan) and subjected to ICC/confocal imaging to detect assembled fibrillar FN (L8 antibody, red). The decrease in assembled FN (L8- red) around the epithelial periphery relative to negative control–treated SMGs and aberrant accumulation of FN (cyan) observed in the interior of the epithelium in LIMK 2 siRNA–treated glands is rescued with simultaneous addition of cofilin and p25 siRNA. Scale, 20 μm.
Because LIMK-mediated signaling resulted in FN assembly and deposition to promote cleft elongation, we investigated whether restoration of cytoskeletal dynamics would stimulate FN fibrillogenesis in the cleft region. We labeled the basement membrane of epithelial rudiments under rescue conditions with trace amounts of Alexa 647–purified human plasma FN and immunostained them with L8 monoclonal antibody. Rescue of LIMK inhibition with cofilin siRNA (Supplemental Figure S6B) or LIMK KD with both p25 and cofilin siRNAs led to basally localized deposition of FN matrix in the clefts of the rescued glands (Figure 10B). In summary, these results confirm that maintenance of cytoskeletal dynamics through cofilin and TPPP/p25 is essential for controlling cleft formation in mouse embryonic salivary glands. LIMK-mediated effects on cytoskeletal stability are indispensable not only for maintaining the structural integrity of the tissue, but also for transducing “inside-out” integrin signaling, leading to organized assembly of FN and thus facilitating the early steps in salivary gland morphogenesis (Figure 11).
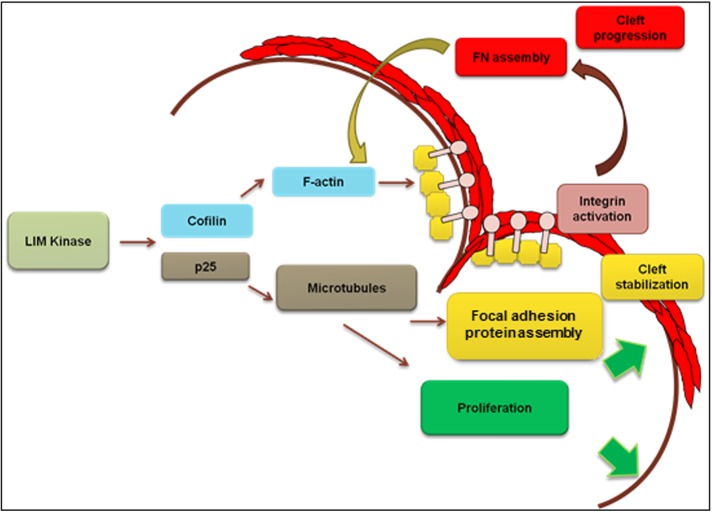
FIGURE 11:
Model for LIMK-mediated regulation of cleft stabilization and cleft progression in mouse E13 salivary glands undergoing branching morphogenesis. Schematic diagram of a proposed model for LIMK-mediated cytoskeletal changes regulating cleft formation: LIMK via cofilin and TPPP/p25 exerts multistep control of cleft formation by affecting focal adhesion assembly and activation and integrin activation to regulate cleft stabilization, and FN assembly and cell proliferation to regulate cleft progression.
DISCUSSION
We here describe a central role for LIMK 1/2 in coordinating cytoskeletal organization in the process of cleft formation during salivary gland branching morphogenesis. We demonstrate that LIMK regulates the early stages of cleft formation—cleft initiation, stabilization and progression—via promotion of F-actin stability. We provide evidence that this function of LIMK requires the actin-severing protein cofilin. Further, we reveal a novel role for the microtubule assembly factor p25 in regulating stabilization and progression of late-stage progressing clefts. Our results thus support a model in which synchronized actin microfilament and microtubule organization are required to exert multistep control on cleft stabilization and progression. Building on our previous work (Daley et al., 2011), we propose that cleft stabilization is not a one-step procedure but contains multiple stabilization steps preceding each event of progression via newly assembled FN. Although cofilin was previously implicated in collective migration of border cells during Drosophila oogenesis (Zhang et al., 2011) and renal branching morphogenesis (Kuure et al., 2010), this is the first report elucidating the role of LIMK and p25 in embryonic organ development.
Our data support the model that cleft formation is highly sensitive to the assembly state of both the actin and microtubule networks. Our apparently paradoxical observation that LIMK inhibition, which causes a decrease in F-actin polymerization, and cofilin KD, which increases F-actin formation, both reduce cleft formation is consistent with an optimal level of actin assembly being required for cleft formation. Given our previous in silico prediction using a local cell-based epithelial model of cleft formation that cleft progression requires an optimal level of actin functionality (Ray et al., 2013), this result is not surprising. Similarly, although LIMK and p25 induced opposing effects on the stability of microtubule network, down-regulation of both proteins perturbed the developmental process. Because the inhibition of LIMK together with p25 and cofilin KD rescued branching morphogenesis, our data support the notion that the assembly state of the cytoskeletal networks is critical for effective branching morphogenesis.
Actin filaments are required for actomyosin contraction, which we previously reported is required for effective cleft progression during branching morphogenesis (Daley et al., 2009). Myosin phosphatase is one factor that is required for regulation of actin contractility, which is done by regulating the activity of nonmuscle myosin type II (Hartshorne et al., 1998). The myosin phosphatase target subunit (MYPT1) of myosin phosphatase was recently shown to mediate a balance between the actin and microtubule cytoskeletons, since it also regulates histone deacetylase 6 (HDAC6) both in fibroblast cells and in developing salivary glands (Joo and Yamada, 2014). This study established MYPT1 as a molecular switch that coordinates levels of actomyosin contractility and microtubule acetylation to promote branching morphogenesis by regulating surface density of α5β1 integrin and hence FN fibrillogenesis (Joo and Yamada, 2014). Of interest, LIMK1, HDAC6, and TPPP1/p25 have been reported to form a trimeric complex that is responsible for increased microtubule acetylation and actin filament destabilization. Under conditions of LIMK1 phosphorylation, the complex disintegrates and allows p25 phosphorylation, leading to microtubule destabilization and F-actin stabilization (Schofield et al., 2014). Although LIMK1 was previously shown to interact directly with p25 through the PDZ domain (Acevedo et al., 2007), it was shown in this study that p25 binds to HDAC6, causing inhibition of its activity. Thus LIMK may regulate microtubule stability through p25 regulation of HDAC6 to control FN fibrillogenesis.
Our observation that LIMK-dependent alterations in the cytoskeleton resulted in reduced assembly and recruitment of FA proteins is consistent with a role for LIMK in cleft stabilization by regulation of focal adhesion complexes. We previously established that focal adhesion complex formation is critical for cleft progression in developing salivary glands (Daley et al., 2011), and our data suggest that LIMK participates in recruitment of focal adhesion proteins and FAK phosphorylation. More than 100 proteins have been implicated in focal adhesion complexes (Schaller, 2001), and the specific complexes formed are dependent on the particular cell type (Zouq et al., 2009). We previously noted that paxillin levels were not significantly affected by inhibition of ROCK signaling during E13 SMG development. Similarly, we noted that paxillin phosphorylation on the Src phosphorylation site (Tyr-118; Sachdev et al., 2009) was not significantly affected by LIMK inhibition, suggesting that paxillin may not be required for cleft formation. LIMK-mediated focal adhesion protein recruitment within the cleft region was associated with activation of integrin β1, as detected with the conformation-dependent antibody 9EG7. LIMK was found to participate in stimulation of “inside-out” integrin β1 activation, which is associated with commencement of FN fibrillogenesis in the clefts (Sakai et al., 2003, Larsen et al., 2006).
Epithelial cell polarization is an important cellular event during branching morphogenesis that may also involve LIMK. LIMK2 activity was previously found to be negatively regulated by the apical par protein, Par3, and Par3 binding to LIMK2 regulated actin turnover during cell polarization (Chen and Macara, 2006). Our results demonstrate that inhibition of LIMK induced a loss of cellular organization in the epithelium, with a loss of outer cuboidal cell organization and a reduction in the area and perimeter of all epithelial cells. Of interest, with LIMK KD we also detected accumulation of apparently extracellular FN within the epithelial bud, reminiscent of aberrant deposition of FN by mispolarized interior epithelial cells induced by ROCK inhibition (Daley et al., 2012). Because our previous work showed that ROCK1-mediated Par1b localization was required to maintain accurate basement membrane deposition (Daley et al., 2012), further investigation of a role for LIMK in epithelial cell polarity during salivary gland branching morphogenesis is warranted.
In this work, we focused on the epithelial functions of LIMK in branching morphogenesis; however, LIMK may have additional roles in the mesenchyme. In fact, we demonstrated a larger inhibition of cleft formation by LIMK KD in intact glands than in epithelial rudiments, suggesting a mesenchymal contribution to cleft formation. In cancer cells, LIMK regulates the expression and activation of MMP 2 and MT1-MMP, affecting ECM remodeling and metastasis (Tapia et al., 2011). Given that MT2-MMP–mediated collagen IV deposition was found to modulate salivary gland branching morphogenesis (Rebustini et al., 2009), LIMK may have an additional role in the regulation of MMP activity during early SMG development to control extracellular matrix and/or basement membrane remodeling. LIMK has known functions in neuronal morphogenesis and differentiation in hippocampal neurons, neuroblastoma cells, and neuronal cell lines (Manetti, 2012). Because parasympathetic innervation is required for effective salivary gland branching morphogenesis (Knox et al., 2010), LIMK1 may be involved in regulation of neuronal morphogenesis during salivary gland development. Cofilin phosphorylation was previously implicated in semaphorin 3A–induced neuronal growth cone collapse (Aizawa et al., 2001). In mouse salivary glands, semaphorin 3A and 3C were found to induce local epithelial signaling to regulate cleft formation via neuropilin 1 (Npn1) without affecting tissue proliferation (Chung et al., 2007). It will be interesting to examine whether LIMK is involved in semaphorin 3A–induced signaling in the SMG and whether this pathway involves parasympathetic innervation. Further research is required to delineate the comprehensive contributions of LIMK to branching morphogenesis in developing salivary glands.
MATERIALS AND METHODS
Submandibular salivary gland ex vivo organ culture and inhibitor assays
Mouse SMGs were harvested from timed-pregnant female mice (strain CD-1; Charles River Laboratories, Troy, NY) at E13, as described previously (Bilgin et al., 2012; Daley et al., 2012; Ray et al., 2013). SMGs were microdissected and cultured as six glands per condition, bright-field images were acquired, and fold changes in bud number were calculated as previously described (Daley et al., 2012; Ray et al., 2013). SMG epithelial rudiments (five glands per condition) were cultured in a gel with final concentration of 6 mg/ml Matrigel (BD Biosciences, San Jose, CA) diluted in DMEM/F12 (Life Technologies, Grand Island, NY) containing 200 ng/ml FGF7 (R&D Systems, Minneapolis, MN). Pharmacological inhibitors were resuspended in DMSO vehicle: BMS-5 (Syn1024, 15 μM; SYNkinase, San Diego, CA) (Ross-Macdonald et al., 2008), brefeldin A (5 μM; EMD Millipore, Billerica, MA), and Taxol (250 nM; EMD Millipore).
siRNA knockdown of LIMK 1, LIMK 2, TPPP/p25, cdc42, and cofilin
The following duplexed siRNAs (predesigned Ambion Silencer Select) were obtained from Life Technologies: LIMK 1, 156129 and s69232; LIMK 2, 156133 and s69236; TPPP/p25, s91408 and s91407; nonmuscle cofilin, Cfl1, s63902; cdc42 siRNA, s63741; and negative control siRNA # 1. SMGs were transfected with 500 nM siRNA using RNAiFect (Qiagen, Valencia, CA) and cultured for 24–48 h, as we reported previously (Daley et al., 2009)
Proliferation and apoptosis assays
For cell proliferation studies, intact E13 SMGs were cultured for 24 h in the presence of inhibitor or vehicle control, and Click-iT Edu (Life Technologies) was used to detect S-phase cells. SMGs were counterstained with SYBR Green I, and proliferation was measured as the ratio of total pixel intensity of EdU-positive cells to that of SYBR Green I staining. Apoptosis was assayed with cleaved caspase-3 to detect apoptotic cells and SYBR Green to label total nuclei. Ratios of EdU/SYBR green and cleaved caspase-3/SYBR green were calculated from confocal stacks spanning the thickness of the entire SMG and graphed in Prism (GraphPad).
Whole-mount immunocytochemistry and fluorescence intensity measurements
Whole-mount immunocytochemistry was performed essentially as described (Daley et al., 2012; Ray et al., 2013). SMGs were imaged either as a stack or at the center of each explant on a Zeiss LSM 510 Meta confocal microscope at 20× (Plan Apo/0.75 numerical aperture [NA]) or 63× (Plan Apo/1.4 NA) magnification. All primary antibodies were used at a 1:100 dilution of the stock solution (unless otherwise noted) with appropriate cyanine dye–conjugated F(ab)2 secondary antibodies (1:100; Jackson ImmunoResearch Laboratories, West Grove, PA). The following antibodies were used: LIMK 1 (Cell Signaling, Danvers, MA), LIMK 2 (clone EP969Y; Abcam, Cambridge, MA), anti–type IV collagen (clone AB769; Millipore), E-cadherin (clone 36, BD Biosciences, San Jose, CA), β-tubulin (clone TUB 2.1; Sigma-Aldrich, St. Louis, MO), cofilin (phospho-Ser-3; Signalway, College Park, MD), talin (clone 8d4; Sigma-Aldrich), vinculin (clone hVIN-1; Sigma-Aldrich), integrin β1 (CD 29; clone EP1041Y; Epitomics, Burlingame, CA), purified rat anti-mouse CD29 (clone 9EG7; BD PharMingen, San Jose, CA), cleaved caspase-3 (Asp-175; Cell Signaling), Cdc 42 (C20; Santa Cruz Biotechnology, Dallas, TX), fibronectin rabbit polyclonal antibody R5836 (1:25), and L8 monoclonal antibody (0.2 mg/ml). SYBR Green I (1:100,000; Life Technologies), Alexa Fluor 488–phalloidin (1:350; Life Technologies), and fluorescent deoxyribonuclease I conjugates (1.5 μM; Molecular Probes, Grand Island, NY) were also used for imaging.
For each image of an immunostained explant, epithelial area was selected in each gland, and total pixel intensity was measured using ImageJ (National Institutes of Health, Bethesda, MD) for the protein markers (F-actin, G-actin, phospho-cofilin, and β-tubulin) and for SYBR Green–stained nuclei. The ratio of pixel intensity of protein markers/SYBR green was calculated from confocal stacks spanning the thickness of the entire SMG and graphed in Prism.
FN labeling
Human plasma fibronectin (Millipore) was conjugated with Alexa Fluor 647 Protein Labeling Kit (Life Technologies) and dialyzed overnight with Tube-o-dialyzer 4 kDa Micro (GBiosciences, St. Louis, MO). Labeled human fibronectin was added at 0.04 μg/ml to live, mesenchyme-free epithelial rudiments for analysis of assembled fibronectin.
Western analysis
Protein assays (Pierce, Rockford, IL) and Western blot analysis (Bio-Rad, Hercules, CA) were performed as previously described (Cantara et al., 2012) with 8 μg of protein loaded/well and protein bands quantified using ImageJ (version 1.46r) with normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Levels of each phosphorylated protein were additionally normalized to total protein levels. Antibodies, their phosphorylated epitope (when relevant), their dilution from stock solution, and their source are as follows: GAPDH (1:10,000; Fitzgerald, Acton, MA), p25/TPPP (clone EPR 3316, 1:1000; Epitomics), phospho–LIMK 1 (Thr-508; 1:500; Abcam), phospho–LIMK 2 (T505; 1:500; Abcam), phospho-cofilin (Ser-3; 77G2, 1:1000; Cell Signaling), cofilin (1:2000; Sigma-Aldrich), phospho-FAK (Tyr-576/577; 1:1000; Cell Signaling), phospho-FAK (1:1000; Tyr-397; Cell Signaling), FAK (1:1000; Cell Signaling), phospho-paxillin (Tyr-118; 1:1000; Cell Signaling), paxillin (1:500; Cell Signaling), phospho–histone H3 (pHH3; Ser-10; 1:1000; Cell Signaling), and histone H3 (1:1000; Cell Signaling).
Statistical analysis
For all studies, either a paired t test or one-way analysis of variance (ANOVA), as appropriate, followed by a Bonferroni posttest from at least three independent experiments was performed using Prism. Error bars represent SEM.
Bright-field time-lapse microscopy and cleft depth analysis
Time-lapse microscopy was performed using a Carl Zeiss microscope essentially as previously described (Daley et al., 2009). Bright-field images were captured every 5 min for 24 h using a Zeiss Cell Observer Z1 fitted with an environmental chamber using AxioVision software (SE64 release 4.9.1; Carl Zeiss) at 5× (Plan Neo, NA 0.16) or 20× (Plan Neo, NA 0.5) magnification. For cleft measurements, TIFF images were imported into MetaVue (version 7.7.5.0; Molecular Devices). Morphometric measurements were acquired using the line tool from calibrated images of 12 clefts from six glands grown under each growth condition.
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health Grants RO1 DE019244 and C06 RR015464. We acknowledge Nimit Dhulekar and Bulent Yener, Department of Computer Science, Rensselaer Polytechnic Institute (Troy, NY), for computational quantification of epithelial cell size and Sharon Sequeira and Deirdre Nelson, University at Albany, State University of New York, for helpful discussions. The total FN and L8 antibodies were generously provided by Kenneth Yamada.
Abbreviations used:
- DMSO
dimethyl sulfoxide
- ECM
extracellular matrix
- FA
focal adhesion
- FAK
focal adhesion kinase
- FN
fibronectin
- HDAC6
histone deacetylase 6
- KD
knockdown
- LIMK
LIM kinase
- MYPT1
myosin phosphatase target subunit 1
- pHH3
phospho histone H3
- siRNA
small interfering RNA
- TPPP
tubulin polymerization-promoting protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-02-0705) on June 25, 2014.
*Present address: Department of Dentistry, University of Pennsylvania School of Dental Medicine, Philadelphia, PA 19104.
REFERENCES
- Acevedo K, Li R, Soo P, Suryadinata R, Sarcevic B, Valova VA, Graham ME, Robinson PJ, Bernard O. The phosphorylation of p25/TPPP by LIM kinase 1 inhibits its ability to assemble microtubules. Exp Cell Res. 2007;313:4091–4106. doi: 10.1016/j.yexcr.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Acevedo K, Moussi N, Li R, Soo P, Bernard O. LIM kinase 2 is widely expressed in all tissues. J Histochem Cytochem. 2006;54(5):487–501. doi: 10.1369/jhc.5C6813.2006. [DOI] [PubMed] [Google Scholar]
- Aizawa H, et al. Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3a-induced growth cone collapse. Nat Neurosci. 2001;4:367–373. doi: 10.1038/86011. [DOI] [PubMed] [Google Scholar]
- Bilgin CC, Ray S, Baydil B, Daley WP, Larsen M, Yener B. Multiscale feature analysis of salivary gland branching morphogenesis. PloS One. 2012;7:e32906. doi: 10.1371/journal.pone.0032906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Shattil SJ, Ginsberg MH. Integrins and actin filaments: reciprocal regulation of cell adhesion and signaling. J Biol Chem. 2000;275:22607–22610. doi: 10.1074/jbc.R900037199. [DOI] [PubMed] [Google Scholar]
- Cantara SI, Soscia DA, Sequeira SJ, Jean-Gilles RP, Castracane J, Larsen M. Selective functionalization of nanofiber scaffolds to regulate salivary gland epithelial cell proliferation and polarity. Biomaterials. 2012;33:8372–8382. doi: 10.1016/j.biomaterials.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Macara IG. Par-3 mediates the inhibition of LIM kinase 2 to regulate cofilin phosphorylation and tight junction assembly. J Cell Biol. 2006;172:671–678. doi: 10.1083/jcb.200510061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Yang TL, Huang HR, Hsu SM, Cheng HJ, Huang PH. Semaphorin signaling facilitates cleft formation in the developing salivary gland. Development. 2007;134:2935–2945. doi: 10.1242/dev.005066. [DOI] [PubMed] [Google Scholar]
- Daley WP, Gervais EM, Centanni SW, Gulfo KM, Nelson DA, Larsen M. ROCK1-directed basement membrane positioning coordinates epithelial tissue polarity. Development. 2012;139:411–422. doi: 10.1242/dev.075366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley WP, Gulfo KM, Sequeira SJ, Larsen M. Identification of a mechanochemical checkpoint and negative feedback loop regulating branching morphogenesis. Dev Biol. 2009;336:169–182. doi: 10.1016/j.ydbio.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley WP, Kohn JM, Larsen M. A focal adhesion protein–based mechanochemical checkpoint regulates cleft progression during branching morphogenesis. Dev Dyn. 2011;240:2069–2083. doi: 10.1002/dvdy.22714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletta VC, Moussi N, Sarmiere PD, Bamburg JR, Bernard O. LIM kinase 1, a key regulator of actin dynamics, is widely expressed in embryonic and adult tissues. Exp Cell Res. 2004;294:392–405. doi: 10.1016/j.yexcr.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Gorovoy M, Niu J, Bernard O, Profirovic J, Minshall R, Neamu R, Voyno-Yasenetskaya T. LIM kinase 1 coordinates microtubule stability and actin polymerization in human endothelial cells. J Biol Chem. 2005;280:26533–26542. doi: 10.1074/jbc.M502921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne DJ, Ito M, Erdo F. Myosin light chain phosphatase: subunit composition, interactions and regulation. J Muscle Res Cell Motil. 1998;19:325–341. doi: 10.1023/a:1005385302064. [DOI] [PubMed] [Google Scholar]
- Heng YW, Lim HH, Mina T, Utomo P, Zhong S, Lim CT, Koh CG. TPPP acts downstream of RhoA–ROCK–LIMK2 to regulate astral microtubule organization and spindle orientation. J Cell Sci. 2012;125:1579–1590. doi: 10.1242/jcs.096818. [DOI] [PubMed] [Google Scholar]
- He L, et al. Modulation of cofilin phosphorylation by inhibition of the Lim family kinases. Bioorg Med Chem Lett. 2012;22:5995–5998. doi: 10.1016/j.bmcl.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Horita Y, Ohashi K, Mukai M, Inoue M, Mizuno K. Suppression of the invasive capacity of rat ascites hepatoma cells by knockdown of Slingshot or LIM kinase. J Biol Chem. 2008;283:6013–6021. doi: 10.1074/jbc.M706538200. [DOI] [PubMed] [Google Scholar]
- Joo EE, Yamada KM. MYPT1 regulates contractility and microtubule acetylation to modulate integrin adhesions and matrix assembly. Nat Commun. 2014:5, 3510. doi: 10.1038/ncomms4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoya Y, Yamashina S. Cellular dynamics of epithelial clefting during branching morphogenesis of the mouse submandibular gland. Dev Dyn. 2010;239:1739–1747. doi: 10.1002/dvdy.22312. [DOI] [PubMed] [Google Scholar]
- Knosp WM, Knox SM, Hoffman MP. Salivary gland organogenesis. Wiley Interdiscip Rev Dev Biol. 2012;1:69–82. doi: 10.1002/wdev.4. [DOI] [PubMed] [Google Scholar]
- Knox SM, Lombaert IMA, Reed X, Vitale-Cross L, Gutkind JS, Hoffman MP. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science. 2010;329:1645–1647. doi: 10.1126/science.1192046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuure S, Cebrian C, Machingo Q, Lu BC, Chi X, Hyink D, D'Agati V, Gurniak C, Witke W, Costantini F. Actin depolymerizing factors cofilin1 and destrin are required for ureteric bud branching morphogenesis. PLoS Genet. 2010;6:e1001176. doi: 10.1371/journal.pgen.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci. 2006;119:3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti F. LIM kinases are attractive targets with many macromolecular partners and only a few small molecule regulators. Med Res Rev. 2012;32:968–998. doi: 10.1002/med.20230. [DOI] [PubMed] [Google Scholar]
- McConnell BV, Koto K, Gutierrez-Hartmann A. Nuclear and cytoplasmic LIMK1 enhances human breast cancer progression. Mol Cancer. 2011;10:75. doi: 10.1186/1476-4598-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima T, Naotsuka M, Horita Y, Sato M, Ohashi K, Mizuno K. LIM-kinase is critical for the mesenchymal-to-amoeboid cell morphological transition in 3D matrices. Biochem Biophys Res Commun. 2010;392:577–581. doi: 10.1016/j.bbrc.2010.01.075. [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Morita T, Nogawa H. Cell proliferation is not required for the initiation of early cleft formation in mouse embryonic submandibular epithelium in vitro. Development. 1987;99:429–437. doi: 10.1242/dev.99.3.429. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci USA. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera T, Sakai T, Hsu JCF, Matsumoto K, Chiorini JA, Yamada KM. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science. 2010;329:562–565. doi: 10.1126/science.1191880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovádi J, Orosz F. An unstructured protein with destructive potential: TPPP/p25 in neurodegeneration. Bioessays. 2009;31:676–686. doi: 10.1002/bies.200900008. [DOI] [PubMed] [Google Scholar]
- Parker KK, Brock AL, Brangwynne C, Mannix RJ, Wang N, Ostuni E, Geisse NA, Adams JC, Whitesides GM, Ingber DE. Directional control of lamellipodia extension by constraining cell shape and orienting cell tractional forces. FASEB J. 2002;16:1195–1204. doi: 10.1096/fj.02-0038com. [DOI] [PubMed] [Google Scholar]
- Patel VN, Rebustini IT, Hoffman MP. Salivary gland branching morphogenesis. Differentiation. 2006;74:349–364. doi: 10.1111/j.1432-0436.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- Ray S, Yuan D, Dhulekar N, Oztan B, Yener B, Larsen M. Cell-based multi-parametric model of cleft progression during submandibular salivary gland branching morphogenesis. PLoS Comput Biol. 2013;9:e1003319. doi: 10.1371/journal.pcbi.1003319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebustini IT, Myers C, Lassiter KS, Surmak A, Szabova L, Holmbeck K, Pedchenko V, Hudson BG, Hoffman MP. MT2-MMP dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev Cell. 2009;17:482–493. doi: 10.1016/j.devcel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P, et al. Identification of a nonkinase target mediating cytotoxicity of novel kinase inhibitors. Mol Cancer Ther. 2008;7:3490–3498. doi: 10.1158/1535-7163.MCT-08-0826. [DOI] [PubMed] [Google Scholar]
- Sachdev S, Bu Y, Gelman IH. Paxillin-Y118 phosphorylation contributes to the control of Src-induced anchorage-independent growth by FAK and adhesion. BMC Cancer. 2009;9:12. doi: 10.1186/1471-2407-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- Schaller MD. Biochemical signals and biological responses elicited by the focal adhesion kinase. Biochim Biophys Acta Mol Cell Res. 2001;1540:1–21. doi: 10.1016/s0167-4889(01)00123-9. [DOI] [PubMed] [Google Scholar]
- Schoenwaelder SM, Burridge K. Bidirectional signaling between the cytoskeleton and integrins. Curr Opin Cell Biol. 1999;11:274–286. doi: 10.1016/s0955-0674(99)80037-4. [DOI] [PubMed] [Google Scholar]
- Schofield AV, Gamell C, Bernard O. LIMK1/TPPP1/HDAC6 is a dual actin and microtubule regulatory complex that promotes drug resistance. Adv Biosci Biotechnol. 2014;5:353. [Google Scholar]
- Scott RW, et al. LIM kinases are required for invasive path generation by tumor and tumor-associated stromal cells. J Cell Biol. 2010;191:169–185. doi: 10.1083/jcb.201002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolich B, Vo M, Buckley S, Plowman G, Papkoff J. Cloning and biochemical characterization of LIMK-2, a protein kinase containing two LIM domains. J Biochem. 1997;121:382–388. doi: 10.1093/oxfordjournals.jbchem.a021599. [DOI] [PubMed] [Google Scholar]
- Spooner BS. Microfilaments, cell shape changes, and morphogenesis of salivary epithelium. Am. Zool. 1973;13:1007–1022. [Google Scholar]
- Spooner BS, Wessels NK. Effects of cytochalasin B upon microfilaments involved in morphogenesis of salivary epithelium. Proc Natl Acad Sci USA. 1970;66:360–364. doi: 10.1073/pnas.66.2.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner BS, Wessels NK. An analysis of salivary gland morphogenesis: role of cytoplasmic microfilaments and microtubules. Dev Biol. 1972;27:38–54. doi: 10.1016/0012-1606(72)90111-x. [DOI] [PubMed] [Google Scholar]
- Sumi T, Matsumoto K, Takai Y, Nakamura T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by Rho- and Cdc42-activated LIM-kinase 2. J Cell Biol. 1999;147:1519–1532. doi: 10.1083/jcb.147.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia T, Ottman R, Chakrabarti R. LIM kinase1 modulates function of membrane type matrix metalloproteinase 1: implication in invasion of prostate cancer cells. Mol Cancer. 2011;10:6. doi: 10.1186/1476-4598-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS. Salivary gland development. Semin Cell Dev Biol. 2007;18:237–244. doi: 10.1016/j.semcdb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta Mol Cell Res. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Wu C, Keivenst VM, O'Toole TE, McDonald JA, Ginsberg MH. Integrin activation and cytoskeletal interaction are essential for the assembly of a fibronectin matrix. Cell. 1995;83:715–724. doi: 10.1016/0092-8674(95)90184-1. [DOI] [PubMed] [Google Scholar]
- Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- Zhang L, Luo J, Wan P, Wu J, Laski F, Chen J. Regulation of cofilin phosphorylation and asymmetry in collective cell migration during morphogenesis. Development. 2011;138:455–464. doi: 10.1242/dev.046870. [DOI] [PubMed] [Google Scholar]
- Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouq NK, Keeble JA, Lindsay J, Valentijn AJ, Zhang L, Mills D, Turner CE, Streuli CH, Gilmore AP. FAK engages multiple pathways to maintain survival of fibroblasts and epithelia–differential roles for paxillin and p130Cas. J Cell Sci. 2009;122:357–367. doi: 10.1242/jcs.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.