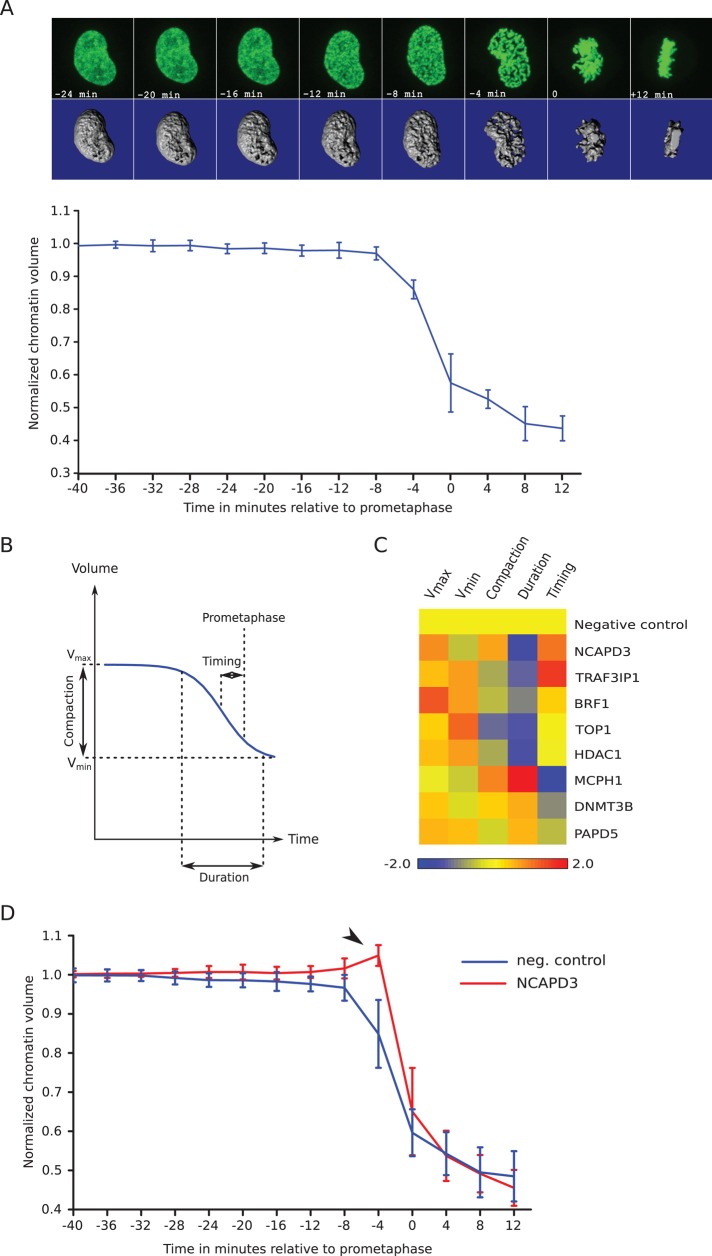
FIGURE 5:
Quantification of chromosome condensation. (A) Evolution of normalized chromatin volume in control cells. Top, maximum intensity projection images (top) and isosurface reconstruction (bottom) of the chromatin from a representative scrambled siRNA–treated control cell at different time points. Occasionally cell rounding in metaphase brings chromatin slightly out of imaging range, resulting in a missing image slice. The missing part is then estimated (see Materials and Methods). The curve represents the average of 10 control cells from four independent experiments. Error bars represent SDs of the means. (B) Definition of chromosome condensation parameters. A sigmoidal decay curve (blue line) is fitted to normalized chromatin volume over time. The fit defines the maximum and minimum volumes, the compaction ratio, and the duration and timing of condensation relative to prometaphase. (C) Heatmap of chromosome condensation parameters in gene-knockdown experiments. The color scale encodes the number of pooled SDs away from the negative control mean. Values smaller than in control are in blue, and values higher than in control are in red. (D) Absence of prophase condensation correlates with chromatin expansion before NEBD. NCAPD3-knockdown cells with almost no prophase (red curve) show transient chromatin decondensation at the time of NEBD (arrowhead), whereas this is never seen in control cells (blue curve). Error bars show SDs of the means (n = 25 for control cells, n = 10 for NCAPD3-knockdown cells).