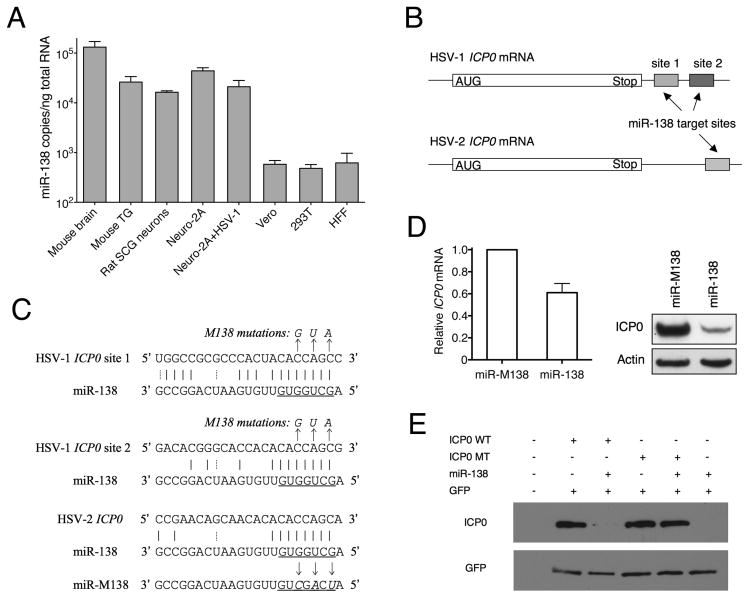
Figure 1.
Neuron-specific miR-138 can target HSV ICP0. (A) qRT-PCR analysis. miR-138 copy numbers normalized to amounts of total RNA in the various tissues and cells indicated below the graph. Neuro-2A + HSV-1; MOI = 50, and 16 hpi. Assays were performed in triplicate with standard deviations shown as error bars. (B) Schematic diagram showing candidate miR-138 target sites in the 3′ UTRs of HSV-1 and HSV-2 ICP0 mRNAs. White boxes indicate the coding sequences. Candidate miR-138 target sites are shown in hatched and grey boxes. (C) Sequences of candidate miR-138 target sites. Below each candidate target site is the sequence of miR-138 (conserved in mouse, rat, monkey and human). Underlined are miR-138 seed sequences. Solid and dashed vertical lines signify Watson-Crick and G-U wobble base pairs, respectively. Arrows point to nucleotide substitutions in mutant forms of target sites and miR-138 with altered bases in italics. (D) miR-138 mimic reduces ICP0 mRNA and protein levels. 293T cells were co-transfected with miR-138 or miR-M138 mimics and an ICP0 WT plasmid. Left, ICP0 mRNA levels normalized to the levels of human GAPDH as measured by qRT-PCR in triplicate. The error bar indicates the standard deviation. Right, ICP0 protein levels analyzed by Western blot in the upper panel and actin in the lower panel as a loading control. (E) miR-138 expressed from a expression plasmid downregulates ICP0 protein expression. 293T cells were mock-transfected or co-transfected with pcDNA-miR-138 plasmid, a WT or M138 ICP0 expression plasmid, and a GFP expression plasmid (control), as indicated above the images. ICP0 (top panel) and GFP (bottom panel) protein levels were assessed using Western blots.