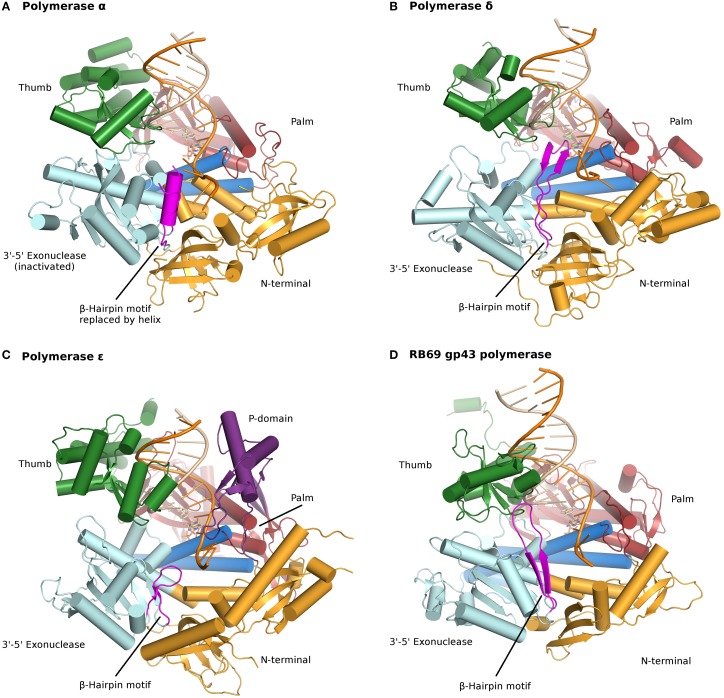
Figure 1.
Ternary complexes of polymerases α, δ, ε, and RB69 gp43 are illustrated from identical orientations for comparison. The thumb (green) and fingers (dark blue) domains grasp the duplex nucleic acid (primer shown in beige, template in orange) against the palm domain (red). The N-terminal domain appears in gold, adjacent to the 3′–5′ exonuclease domain (cyan). (A) Polymerase α (PDBID 4FYD) binds an RNA/DNA hybrid, where the wide, shallow minor groove of A-form DNA is apparent near the thumb. The 3′–5′ exonuclease domain is devoid of activity. A helical region (magenta) in the inactivated exonuclease domain stabilizes the 5′end of the template. (B) Polymerase δ (PDBID 3IAY) harbors a large β hairpin motif (magenta), which is important in switching the primer strand from the polymerase active site to the exonuclease active site in the event of proofreading. (C) Polymerase ε (PDBID 4M8O) wields a unique P-domain (purple), which endows the polymerase with increased processivity. Interestingly, the β hairpin motif is atrophied in pol ε. (D) Conservation of the family B DNA polymerase fold, and domain organization, is evident when the model enzyme from bacteriophage RB69 gp43 (PDBID 2OZS) is viewed along with the three eukaryotic replicative polymerases. The domain delineation for each polymerase is given in Table S1. Figure was made with PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.).