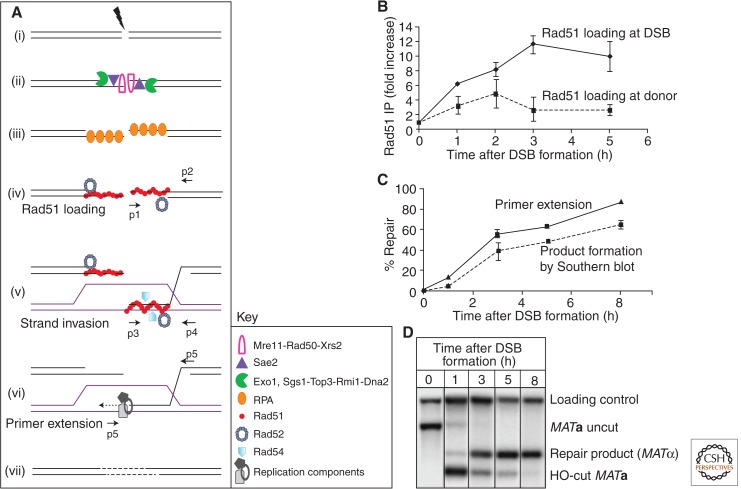
Figure 3.
Key intermediate steps of HR and methods to study them. (A) Key proteins are depicted in sequential early steps in GC in budding yeast. DSB formation (i) is immediately followed by 5′ to 3′ resection (ii). The 3′ tails are stabilized by RPA (iii), which is then replaced with Rad51 recombinase with assistance from accessory proteins like Rad52 (iv). Once the homologous donor is found, strand invasion occurs, resulting in formation of a D-loop (v) by displacement of the identical strand and base pairing with the complementary strand at the donor. (vi) Various components of the replication machinery assemble to start copying from the donor template. (vii) The break is sealed. Small arrows denote positions of PCR primer pairs used to analyze intermediate steps, shown on the right for MAT switching in S. cerevisiae. (B) Recruitment of Rad51 at DSB site (the MATa locus) by chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR) using primers p1 and p2 (solid line). Rad51 binding to the donor template (budding yeast HMLα locus) using primers p3 and p4 for qPCR (dotted line). Error bars indicate standard error of the mean. (C) The initiation of new DNA synthesis by primer extension is detected by using PCR primers p5 and p6, which amplifies a unique fragment once new DNA synthesis has been initiated (solid line). A dotted line shows quantitative densitometric analysis of Southern blot (D) that follows GC progression in budding yeast as MATa switches to MATα. Data from the investigators (A Mehta and J Haber, unpubl.).