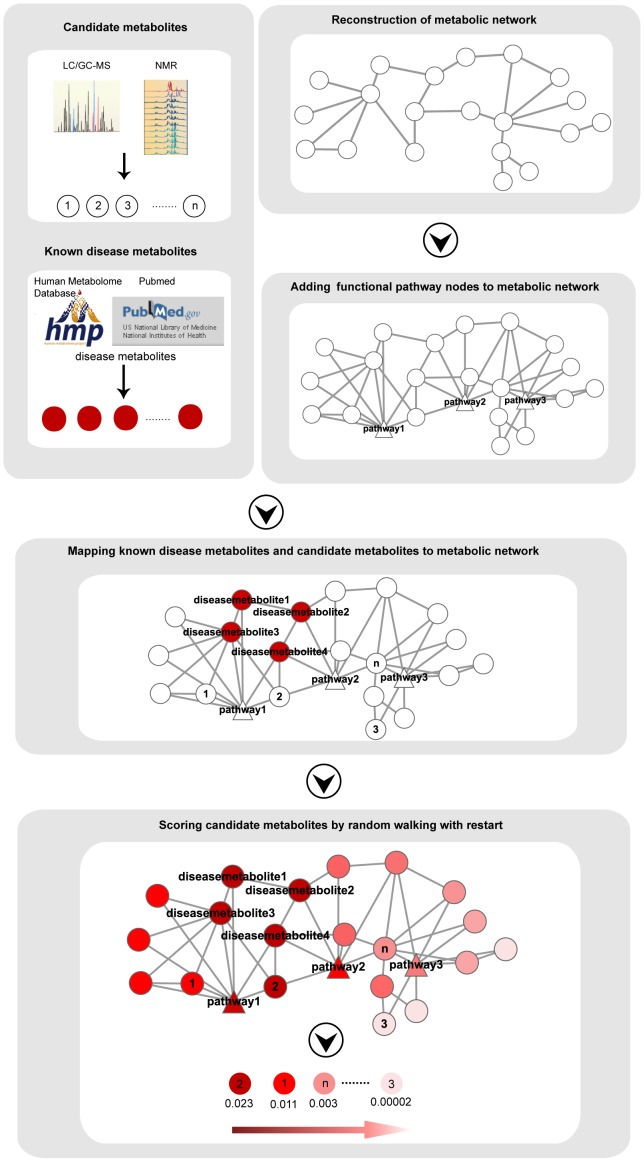
Figure 1. Schematic of the PROFANCY. We firstly reconstruct metabolic networks based on the structure data from KEGG or EHMN database and add functional pathway nodes in this metabolic network.
We then map the known disease metabolites (seed nodes) and candidate metabolites into the above network. After that, we extend random walk with restart (RWR) method to this network. Finally, we can rank the candidate metabolites according to the steady probability of RWR.