Summary
Background
Accurate and reproducible measurement of expression of pro-inflammatory cytokines in colonic biopsies from patients with ulcerative colitis (UC) is essential for proof-of-concept and mechanism-of-action studies. Few studies have rigorously established the number of biopsies required for accurate and reproducible biomarker measurements.
Aim
To validate methods for measuring changes in gene expression in colonic biopsy samples.
Methods
Twelve colonic biopsies were obtained from each of 6 healthy controls, 6 patients with inactive UC, and 7 patients with active UC. Mayo endoscopic scores were used as a clinical reference standard. Quantitative PCR was used to assess mRNA expression of eight known inflammatory genes. The power to detect a reduction in gene expression in active versus inactive UC was calculated using a linear mixed effect model.
Results
mRNA analysis of colonic biopsies is a sensitive and feasible approach for measuring inflammatory gene expression in colonic biopsies. Inflammatory biomarkers correlate with Mayo endoscopic subscores for each colonic region. For most genes, 3 rectal biopsies from 2–4 patients are required to detect changes in gene expression corresponding to active versus inactive UC to achieve a power of 80% with an alpha of 0.05.
Conclusions
Our data suggest that systematic measurement of inflammatory biomarkers at the mRNA level can be a valuable tool for hypothesis testing and assessment of clinical activity and response to therapy in ulcerative colitis.
Keywords: ulcerative colitis, gene expression, coefficient of variation, sampling error, mRNA, molecular targets, biomarkers, power
INTRODUCTION
Ulcerative colitis (UC) is an inflammatory disease involving an aberrant immune response in genetically predisposed individuals with complex environmental and microbial contributions.1 Current treatment options are limited to aminosalicylates, steroids, immunosuppressive agents such as azathioprine, and tumor necrosis factor-alpha (TNFα) antagonists.2 As the understanding of the pathogenesis of UC advances, novel medications targeting different signaling pathways have emerged as potential therapies.3 The currently utilized approach of evaluating candidate drugs with clinical and endoscopic endpoints in early phase 1 and 2 clinical trials requires large numbers of patients for each drug, and is inefficient and unsustainable. Evolution to proof-of-concept studies in which objective, quantitative changes in intestinal inflammation in response to pharmacologic therapy are measured could potentially reduce the number of patients required to evaluate novel therapies.
The ability to measure mRNA expression of genes for pro-inflammatory cytokines, chemokines, and other inflammatory mediators from colonic biopsies of patients with UC in an accurate, precise, and reproducible manner is essential for implementation of proof-of-concept and mechanism-of-action studies. Given the lack of predictive serologic biomarkers of inflammation, assessment of inflammation from colonic biopsies is appealing given their accessibility in the context of clinical trials that include serial endoscopic evaluation of colon inflammation to assess mucosal healing and response to treatment.4
Gene expression studies in patients with inflammatory bowel disease (IBD) have shown that inflammatory cytokines, chemokines, and other inflammatory mediators are expressed in areas of active and inactive disease, and correlate with endoscopic disease activity.5–9 Microarray and confirmatory quantitative PCR (qPCR) techniques have been used to identify genes that are expressed at higher levels in the setting of active disease and normalize with therapy, providing further insights into the signaling pathways aberrantly regulated in IBD.10 It has been recently demonstrated that changes in gene expression signatures correlate with clinical, endoscopic, and histologic responses to treatment and may represent promising sensitive and specific biomarkers of response to therapy.11 Since gene expression in colonic biopsies from healthy individuals varies by anatomic region of the colon,12 measuring gene expression in a single colonic biopsy may yield variable results that are not reproducible.
Little data exist on the numbers of patients or biopsies per patient required to generate accurate, precise, and reproducible results. Existing studies have not systematically evaluated colonic biopsies from different regions of the colon, or even within the same region, to determine the sensitivity of various numbers of biopsies to accurately detect changes in gene expression. We sought to determine the number of colonic biopsies that need to be obtained from a given region of the colon in order to precisely measure gene expression, such that gene expression level could be reliably compared at different times. We used methodology previously developed for mechanism-of-action studies in patients with rheumatoid arthritis (RA) to measure gene expression with quantitative PCR using cell-based standards from mitogen-stimulated peripheral mononuclear cells.13 In our study, we sought to evaluate the precision, accuracy, and reproducibility of measuring gene expression in colonic biopsies from patients with UC to validate methods applicable to early proof-of-concept clinical trials.
MATERIAL AND METHODS
Patient Selection
Patients with UC and healthy individuals who were undergoing colonoscopy as part of routine medical care at the University of California San Diego were enrolled in the study. Patients enrolled had an established diagnosis of UC, had not undergone colectomy, and were ≥18 years old. A Mayo Clinic endoscopic subscore was determined for each region of the colon (ascending, transverse, descending, and rectosigmoid), and each region was then classified as having active or inactive disease. Each participant was classified as having active or inactive disease based on the endoscopic disease activity by a single experienced endoscopist (WJS). The clinical disease activity was calculated for each patient with the Mayo Clinic score for assessment of ulcerative colitis that includes measures of stool frequency, rectal bleeding, endoscopic findings, and global health assessment.14 The protocol was approved by the institutional review board at UCSD, and all patients gave written informed consent.
Colonic Biopsies
Three biopsies were obtained with cold biopsy forceps from each of 4 colonic regions (ascending, transverse, descending, and rectosigmoid) for a total of 12 colonic biopsies from each individual. If a colonic segment was classified as having active disease, the colonic biopsies were obtained from areas with the most evident erythema. Each colonic biopsy was immediately placed into RNA Stat-60 (Tel-Test, Friendswood, TX, USA) and then snap-frozen in liquid nitrogen and stored at −80C. Assays were performed within six months of sample acquisition.
RNA isolation and cDNA synthesis
Each biopsy was homogenized separately on ice in RNA Stat-60 using chilled 1ml Kontes-Duall tissue grinders until only insoluble fibrous materials remained. A crude RNA extract was obtained after addition of chloroform and centrifugation. The resultant aqueous phase was mixed with an equal volume of 70% ethanol and loaded onto RNeasy Lipid Tissue Mini columns (Qiagen, Germantown, MD). Manufacturer protocol was then followed and the RNA was eluted in 40 μl of TE buffer. RNA was quantified using Ribo-green reagent (Life Technologies, Carlsbad, CA). A total of 250ng RNA was used for the reverse transcription reaction (TaqMan Reverse Transcription Reagents, Life Technologies, Carlsbad, CA). Reagents for the Q-PCR including primers for human GAPDH, interleukin (IL)-1β, IL-6, IL-8, TNFα, interferon gamma-induced protein (IP)-10, C-X-C motif chemokine (CXCL)-13, matrix metalloproteinase (MMP)-3, and S100 calcium binding protein A8 (S100A8) were obtained from Life Technologies (Carlsbad, CA). Q-PCR, standards, and normalization with GAPDH were carried out as described by Boyle and colleagues.13 Eight representative genes were selected that encode inflammatory markers implicated in the pathogenesis of IBD and found to be elevated in prior studies.9, 12, 15–17 The inter-assay reproducibility based on the coefficient of variation was 4 – 6% for the eight selected genes. IL-1β, IL-6, IL-8, and TNFα are pro-inflammatory cytokines that are induced through activation of nuclear factor-κB (NF-κB) and mitogen activated protein (MAP) kinases implicated in the pathogenesis of autoimmunity.9, 10, 15 Upon induction by interferon activation, IP-10 acts as an inflammatory cell chemoattractant,17 and CXCL-13 or B lymphocyte chemoattractant (BLC) is a potent chemoattractant critical in initiating formation of lymphoid follicles.18 MMP-3 acts as an inflammatory mediator, degrading extracellular matrix in inflammatory conditions.12, 19 S100A8 is a calcium-binding protein expressed by neutrophils and is a component of calprotectin, a fecal biomarker for inflammation.16, 20
Data Analysis
To evaluate the relationship between gene expression and either endoscopic or clinical disease activity scores, linear mixed effects models were fit to control for repeated biopsies taken within a region for each subject. We reported the estimated regression coefficient associated with the linear effect of the Mayo score and the endoscopic sub-score. Significance was determined with an approximate F-test using the Kenward-Roger approach.21 To compare the gene expression between regions of active versus inactive UC, a cell means mixed model were fit to control for repeated biopsies within a region. The coefficient of variation was calculated for each inflammatory gene biomarker using repeat measures in patients with UC as well as in healthy controls. For the power analyses, we used a simulation based approach using estimates derived from a linear mixed model for the log of relative gene expression with repeat measures within each region of the colon. P-values of <0.05 were considered statistically significant. All analyses were done in the open source statistical language R22 with the packages ‘lme4’23 and ‘pbkrtest.’24
RESULTS
Our biopsy collection strategy was designed to evaluate variation in biopsies taken within a region as well as variation between regions. Colonic biopsies were obtained from 6 healthy controls and 13 patients with UC, 6 with inactive and 7 with active disease. The healthy controls, who were patients undergoing colonoscopy for colon cancer screening or surveillance, had a mean age of 61 years. The mean age of the patients with active and inactive disease was 50 and 40 years old, respectively (Table 1). At the time of colonoscopy, 57% of patients with active disease had left-sided colitis, and 43% had pan-colitis. Forty-three percent of patients with active and 66% of patients with inactive disease were men. Of the patients with active disease, 29% were receiving steroids, whereas none of the patients with inactive disease were receiving steroids. Twenty-nine percent of patients with active disease as compared to 33% of patients with inactive disease were receiving TNFα antagonists. Fourteen percent of patients with active disease and 50% of patients with inactive disease were receiving immunosuppressive therapy with 6-mercaptopurine, azathioprine, or methotrexate. Seventy-one percent of patients with active disease were receiving mesalamine derivatives as compared with 33% of patients with inactive disease.
Table 1.
Baseline Characteristics of Patients with Ulcerative Colitis
| Active (n= 7) | Inactive (n=6) | P-value | |
|---|---|---|---|
|
| |||
| Age (years +/− S.D.) | 50.0 +/− 20.4 | 40.2 +/− 13.0 | NS |
|
| |||
| % Male | 43% | 66% | NS |
|
| |||
| TNFα Antagonists Use, % | 29% | 33% | NS |
|
| |||
| Steroid Use, % | 29% | 0% | NS |
|
| |||
| Immunosuppressive Use, % | 14% | 50% | NS |
|
| |||
| Mayo Clinic Score (mean +/− S.D.) | 6.3 +/− 2.1 | 0.3 +/− 0.5 | 0.003 |
|
| |||
| Mayo Clinic Endoscopy Score (mean +/− S.D.) | 2.1 +/− 0.9 | 0.2 +/− 0.9 | < 0.001 |
|
| |||
| Extent of Colitis | |||
| Left-Sided | 57% | 0% | -- |
| Pan-Colitis | 43% | 0% | -- |
S.D. indicates standard deviation.
The mean Mayo Clinic score was significantly greater for patients with active as compared with inactive disease (Table 1). The mean Mayo Clinic endoscopic score was significantly greater in patients with active (2.1, SD 0.9) as compared with inactive (0.2, SE 0.9, p<0.001) disease. Histologic evaluation of biopsies revealed active inflammation in 86% of patients in the patients classified as having endoscopically active disease compared to 17% of patients classified as having inactive disease.
Correlation of Endoscopic Activity and Mayo Score with Gene Expression
To evaluate the validity of inflammatory gene expression measurements, we evaluated the relationship between gene expression and the Mayo endoscopic subscore using a linear mixed effects model, controlling for regional variation and repeated measures within each subject. Expression of all inflammatory genes measured was significantly associated with the Mayo endoscopic subscore for each colonic region where the coefficient is the estimated increase in log of gene expression for an increase in the endoscopic score (p-value of <0.05). Based on the coefficient value, IL-8 and MMP-3 had the strongest association with the endoscopic subscore, a relatively objective measure of clinical disease activity (Table 2). Using a similar linear regression model, the association between inflammatory gene expression and clinical disease activity, as measured by the Mayo Clinic score, was evaluated, showing a significant association with expression of IL-1β, IL-6, IL-8, MMP-3, IP-10, and S100A8 (p-values <0.05). In contrast, we did not find a significant association between the Mayo Clinic score and TNFα or CXCL-13 expression (p-values of 0.14 and 0.36, respectively). Furthermore, the Mayo Clinic score was not found to have a significant relationship with endoscopic score (Spearman’s was 0.23 with p-value of 0.45), supporting the notion that the Mayo Clinic score, which is heavily driven by subjective patient symptoms, may not accurately predict severity of intestinal inflammation.25, 26
Table 2.
Association Between Inflammatory Gene Expression and Endoscopic Sub-score or Mayo Score
| Inflammatory Gene | Endoscopic Sub-Score Coefficient | Mayo Score Coefficient |
|---|---|---|
| IL-1β | 0.97* | 0.29* |
| IL-6 | 1.25* | 0.40* |
| IL-8 | 1.50* | 0.53* |
| TNFα | 0.58* | 0.11 |
| IP-10 | 0.87* | 0.24* |
| CXCL-13 | 0.66* | 0.10 |
| MMP-3 | 1.79* | 0.50* |
| S100A8 | 1.20* | 0.54* |
Linear Regression was used to calculate the regression coefficient associated with the effect of endoscopic subscore or Mayo score on gene expression with correction for colonic region. Significance was determined by Kenward-Roger F. P value less than 0.05 was considered significant and is indicated by *. Coefficient represents β2 coefficient from the linear regression where it is the estimated increase in the log of the inflammatory gene expression for one unit increase in composite endoscopic subscore or Mayo score.
Comparison of Gene Expression in Active and Inactive UC
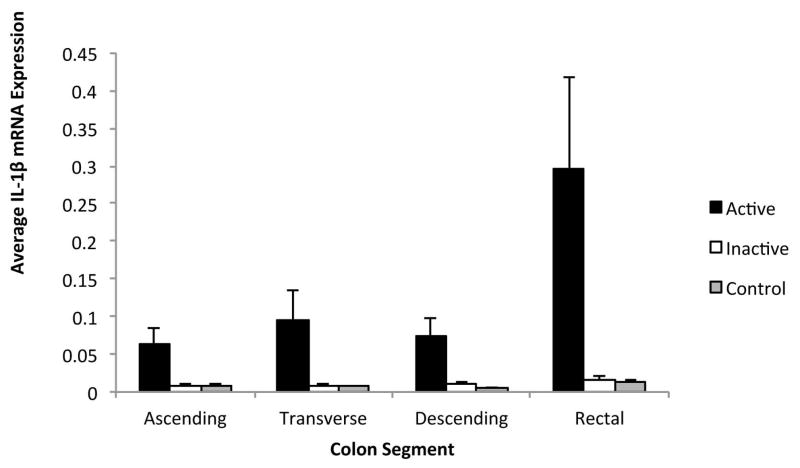
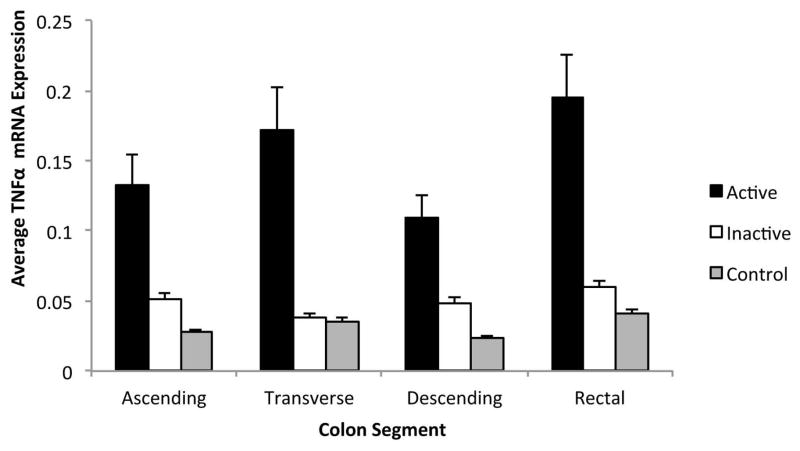
Inflammatory gene expression was compared between active and inactive UC regions using a cell means mixed model. When comparing gene expression from individual colonic regions, the rectal mucosa expression of IL-1β, IL-6, IL-8, MMP-3, IP-10, and S100A8 mRNA was significantly greater in active as compared to inactive colitis. In the proximal colonic regions, expression of IL-1β, IL-8, MMP-3, and S100A8 mRNA was also significantly greater in active as compared to inactive colitis (Figure 1). In contrast, CXCL-13 mRNA expression did not correlate with disease activity in any region, and TNFα mRNA expression was only significantly greater in active as compared to inactive colitis in the transverse colon (Figure 2). These results suggest that specific inflammatory markers, such as IL-1β, IL-8, MMP-3, and S100A8, correlate with disease activity in UC and may serve as the best markers for disease activity.
Figure 1. Comparison of Relative IL-1β Gene Expression by Colonic Region.
Active Ulcerative Colitis, Inactive Ulcerative Colitis, and Controls. Relative IL-1β gene expression units were averaged for patients with active UC, inactive UC, and healthy controls by colonic regions. Gene expression from inactive colonic regions in patients with active UC were excluded. Standard error shown.
Figure 2. Comparison of Relative TNF-α Gene Expression by Colonic Region.
Active Ulcerative Colitis, Inactive Ulcerative Colitis, and Controls. Relative TNF-α gene expression units were averaged for patients with active UC, inactive UC, and healthy controls by colonic region. Gene expression from inactive colonic regions in patients with active UC were excluded. Standard error shown.
Ulcerative Colitis Coefficient of Variation Analysis
In colonic biopsies obtained from individuals with UC, there was significant variability in the coefficient of variation that differed based on the individual inflammatory gene analyzed and the region of the colon sampled. Specifically, the coefficient of variation for TNFα expression from 3 colonic biopsies sampled from a colonic region was relatively low, ranging from 31% in the rectum, 40% in the descending colon, 28% in the transverse colon, to 30% in the ascending colon. Thus, 2 biopsies from the rectum were needed to reduce the standard error for TNFα to less than 25%. In contrast, the expression of CXCL-13 was most variable with the highest coefficient of variation that ranged from 58% in the rectum to 80% in the transverse and descending colon (Table 3). To reduce the standard error to less than 25%, 11 colonic biopsies would need to be obtained from the descending colon. With the exception of the most variably expressed gene CXCL-13, 2–4 rectal biopsies for the remainder of the genes studied would reproducibly reduce the standard error to less than 25% (Table 4). A standard error of 25% was selected a priori as a benchmark based on FDA guidelines for industry bioanalytical method validation.27 Analyses were performed to examine the number of colonic biopsies needed to reduce sampling error to more restrictive values of 10% or 5% (Supplementary Table 1 and 2). These analyses demonstrated, however, that the number of colonic biopsies required from an individual patient to reduce the sampling error to 10% or lower was not feasible. Similarly, inflammatory gene expression in healthy controls varied by gene tested and region of the colon sampled. For example, the variability in TNFα expression appears to be similar for biopsies from each colonic region in healthy controls and patients with UC. However, other genes like CXCL-13 and MMP-3 had higher coefficients of variation that may have been related to lower absolute expression of inflammatory genes (Table 5).
Table 3.
Coefficient of Variation (CV) for Each Inflammatory Gene in a Colonic Region For Patients with Ulcerative Colitis
| Inflammatory Gene | Ascending Colon | Transverse Colon | Descending Colon | Rectum |
|---|---|---|---|---|
| IL-1β | 37% | 26% | 32% | 35% |
| TNFα | 30% | 28% | 40% | 31% |
| IL-6 | 53% | 47% | 56% | 37% |
| IL-8 | 54% | 39% | 43% | 49% |
| MMP-3 | 56% | 40% | 44% | 33% |
| S100A8 | 44% | 35% | 46% | 39% |
| IP-10 | 51% | 29% | 48% | 34% |
| CXCL-13 | 68% | 80% | 80% | 58% |
The coefficient of variation (CV) was calculated for each gene for biopsies from each region of the colon in individuals with UC. The coefficient of variation was different for each inflammatory gene and differed based on region of the colon that was sampled.
Table 4.
Number of Rectal Biopsies from Patients with Ulcerative Colitis Required to Reduce Standard Error to <25%
| Inflammatory Gene | Number of Biopsies to Reduce Std Error to <25% |
|---|---|
| TNFα | 2 |
| IL-1β | 2 |
| IP-10 | 2 |
| MMP-3 | 2 |
| IL-6 | 3 |
| S100A8 | 3 |
| IL-8 | 4 |
| CXCL-13 | 6 |
The coefficient of variation (CV) was calculated for each gene from rectal biopsies in individuals with UC. The number of biopsies needed to reduce standard error to less than 25% was subsequently calculated.
Table 5.
Coefficient of Variation (CV) for Each Inflammatory Gene in a Colonic Region For Healthy Controls
| Inflammatory Gene | Ascending Colon | Transverse Colon | Descending Colon | Rectum |
|---|---|---|---|---|
| IL-1β | 40% | 48% | 50% | 29% |
| TNFα | 40% | 41% | 29% | 42% |
| IL-6 | 50% | 54% | 42% | 55% |
| IL-8 | 66% | 71% | 40% | 40% |
| MMP-3 | 92% | 88% | 69% | 85% |
| S100A8 | 56% | 49% | 64% | 64% |
| IP-10 | 52% | 59% | 44% | 39% |
| CXCL-13 | 91% | 46% | 92% | 53% |
The coefficient of variation (CV) was calculated for each gene for biopsies from each region of the colon in healthy controls. The coefficient of variation was different for each inflammatory gene and differed based on the region of the colon that was sampled.
Mixed Models for Power Analyses
We next tested the applicability of these results to hypothetical future clinical drug trials in which colonic biopsies would be obtained prior to and following treatment. Using two distinct approaches, we asked whether our methodology could detect a response to therapy. In the first approach, we reasoned that a significant response to treatment might result in a hypothetical two-fold reduction in inflammatory gene expression. We therefore calculated the power to detect such a two-fold reduction in inflammatory gene expression, using a linear mixed effect model with repeat measures for each region of the colon. For the majority of the inflammatory genes (IL-1β, IL-8, TNFα, IP-10, MMP-3, and S100A8), three rectal biopsies from 2–4 patients would be the minimum number of biopsies obtained before and after treatment to detect a two-fold change in expression from a given region in the colon with 80% power (Table 6). If a power of 90% were desired to reduce the risk of a type II error, 4–6 patients would be needed to detect changes in the majority of the inflammatory genes (Supplemental Table 3). For IL-6 and CXCL-13, additional subjects or biopsies would be needed to maintain power for studies. Six biopsies from 4 patients for IL-6 and 6 biopsies from 7 patients for CXCL-13 would be needed to obtain a power of 80% in the rectum (Table 6).
Table 6.
Number of UC Patients and Number of Rectal Biopsies Required to Detect a Two-Fold Reduction in Gene Expression
| Number of Patients | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Number of Biopsies | IL-1β | IL-6 | IL-8 | TNFα | IP-10 | CXCL-13 | MMP3 | S100A8 |
| 3 | 3 | 7 | 4 | 2 | 4 | >16 | 4 | 4 |
| 6 | 2 | 4 | 2 | 2 | 2 | 7 | 3 | 3 |
| 9 | 2 | 3 | 2 | 2 | 2 | 6 | 2 | 2 |
Using a linear mixed effect model with repeated measures, the power to detect twofold reduction in inflammatory gene expression from rectal biopsies was calculated. With a fixed number of biopsies before and after treatment, the minimum number of UC patients needed to obtain a power of 80% is reported.
In the second approach, we reasoned that a clinically significant response to therapy might result in a reduction in inflammatory gene expression that resembled that between active versus inactive disease. We therefore used a second linear mixed effect model with repeat measures to determine the power to detect changes in inflammatory gene expression between active versus inactive disease. The relative reduction in inflammatory gene expression ranged from 0.003 to 1.02 (Supplemental Table 4). In the rectum, a total of 3 biopsies pre- and post-treatment would be needed from 2 patients in order to have a power of 80% or up to 90% with alpha of 0.05 for all of the genes except CXCL-13 (Table 7 and Supplemental Table 5). For the majority of the genes, including IL-1β, IL-8, IP-10, MMP-3, and S100A8, 3 biopsies from any region of the colon would be needed from 2–5 patients to maintain statistical power. For the inflammatory genes CXCL-13 and IL-6, in contrast, distinguishing active from inactive disease required larger numbers of colonic biopsies. Moreover, in certain proximal regions, active and inactive disease were indistinguishable using CXCL-13 and IL-6, no matter how many biopsies were taken. Together our data show that carefully selected inflammatory genes, such as IL-1β, IL-8, IP-10, MMP-3, and S100A8, can potentially be used to detect mRNA changes in response to treatment.
Table 7.
Number of UC Patients and Number of Rectal Biopsies Required to Detect Differences in Gene Expression Corresponding to Active versus Inactive Disease
| Number of Patients | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Number of Biopsies | IL-1β | IL-6 | IL-8 | TNFα | IP-10 | CXCL-13 | MMP3 | S100A8 |
| 3 | 2 | 2 | 2 | 2 | 2 | 6 | 2 | 2 |
| 6 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 2 |
| 9 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
Using a linear mixed effect model with repeated measures, the power to detect changes in inflammatory gene expression in rectal biopsies corresponding to active versus inactive UC was calculated. With a fixed number of biopsies before and after treatment, the minimum number of UC patients needed to obtain a power of 80% is reported.
DISCUSSION
Our study systemically evaluates the variation in measuring inflammatory gene mRNA expression and the sensitivity to detect changes in gene expression in a rigorous manner that has not been previously reported. Prior studies have analyzed a single colonic biopsy from a single given area of active or inactive disease, making the assumption that gene expression can be accurately measured from a single sample.28 However, our results suggest that a single sample is insufficient to characterize colonic gene expression and multiple biopsies must be obtained and evaluated in order to reduce sampling error and obtain precise and reproducible results.
There is no accepted standard for replicate sampling in a heterogeneous tissue such as inflamed colon. The technical replicate CV in our quantitative PCR assay ranged from 4 to 6% for all analytes, and thus were all within FDA guidelines for assay variation. To account for tissue variation in colonic sampling, we utilized a standard error of 25%, the least restrictive variation for a single sample assay from FDA guidelines.27 Although the guidelines were intended for ligand binding assays, we used them as a reference for performing power calculations. In effect, we determined the number of biopsies needed to provide sampling reliability equivalent to a single measure of a homogenous tissue.
Recent studies demonstrate that assessment of endoscopic disease activity may be superior to assessment of clinical disease activity (as measured by the Mayo score), in predicting response to therapy.25 In support of this concept, our results indicate that inflammatory gene expression correlates better with endoscopic disease activity than with the Mayo score. These findings suggest that inflammatory markers could be incorporated into clinical trials in addition to other objective measures, such as endoscopic scores, to more fully evaluate response to treatment.
While there is variation in gene expression that depends on the specific region of the colon sampled, the coefficient of variation for the rectum was generally as low as, if not lower than, other regions. For the majority of inflammatory markers, an average of 3 biopsies reduced the standard error to less than 25%, a benchmark for variability in standardized assays. Three to four colonic biopsies from a given segment should be obtained to effectively minimize the variation and obtain precise and reproducible gene expression measurements from a colonic biopsy. Although the study was not specifically powered to detect differences in regional expression of inflammatory genes, differences in inflammatory gene expression between active and inactive UC appeared to be greatest in rectal biopsies. This finding might have been predicted based on the endoscopic appearance of contiguous active UC. Together these findings support the use of flexible sigmoidoscopy with biopsies of the rectum to monitor disease activity and evaluate response to therapies, as is often done in clinical practice. Proximal colonic biopsies did not appear to add significant additional information in distinguishing active and inactive UC.
Our systematic observations of colonic gene expression in UC support a model of hypothesis testing at the site of disease that includes molecular biomarkers closely associated with therapeutic targets. An analogous approach has been previously used to evaluate response to therapy in proof-of-concept and mechanisms-of-action clinical trials in rheumatoid arthritis with serial synovial biopsies.29, 30 By determining variation in gene expression in a systematic manner, we were able to determine the minimum colonic sampling requirements for UC. Moreover, we were able to define the number of patients that needed to be enrolled in a clinical trial and the number of colonic biopsies per patient needed before and after treatment in order to power a study to detect significant changes in gene expression.
In conclusion, molecular biomarker analysis in UC is a feasible, high-yield approach employing highly sensitive, high dynamic range assays. Active and inactive UC can be distinguished with precision using quantitative and objective measures. Three biopsies from 2–4 patients with UC should provide adequate power for proof of concept or mechanism of action studies with novel medications. Our results suggest that broadly defined biomarkers of colonic inflammation can be used to detect a clinical response, potentially with fewer patients than would be needed using conventional clinical, or even endoscopic, endpoints. Thus, gene expression studies using colonic biopsies appear to be an appealing and feasible method to measure quantitative response to therapy and interrogate the mechanism of action in the target tissue of interest at the precise site of mRNA transcription, thereby proving whether a drug is having the desired effect.
Supplementary Material
Acknowledgments
We thank Elizabeth Evans, Marianne Fahmy, Suresh Pola, Preet Bagi, and the endoscopy staff at UCSD Thornton Hospital for their assistance in collecting colonic samples, and the UCSD Clinical Translational Research Institute for their assistance with this project.
Footnotes
Author contributions: Boland, Chang, Sandborn, Boyle, Firestein contributed to the conception, design, and/or oversight of the study. Boland, Pola, Zhang, Copur-Dahi, Proudfoot, Hillman, Sandborn contributed to the collection, analysis, and/or interpretation of data. Boland, Chang, and Sandborn drafted the manuscript. All authors revised the manuscript for important intellectual content and approved the final version of the manuscript.
Declaration of personal interests: None
Declaration of funding interests: This work was supported by US National Institutes of Health (DK093507 to J. Chang) and the UCSD Digestive Diseases Research Development Center Grant DK80506. B. Boland was supported by NIH grant T32 DK07202. B. Zhang was supported by grant 1TLRR03197 from the NIH National Center for Research Resources.
References
- 1.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Ordas I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380(9853):1606–19. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 3.Sandborn WJ. The future of inflammatory bowel disease therapy: where do we go from here? Dig Dis. 2012;30 (Suppl 3):140–4. doi: 10.1159/000342742. [DOI] [PubMed] [Google Scholar]
- 4.Colombel JF, Rutgeerts P, Reinisch W, et al. Early mucosal healing with infliximab is associated with improved long-term clinical outcomes in ulcerative colitis. Gastroenterol. 2011;141(4):1194–201. doi: 10.1053/j.gastro.2011.06.054. [DOI] [PubMed] [Google Scholar]
- 5.Akazawa A, Sakaida I, Higaki S, Kubo Y, Uchida K, Okita K. Increased expression of tumor necrosis factor-alpha messenger RNA in the intestinal mucosa of inflammatory bowel disease, particularly in patients with disease in the inactive phase. Journal of gastroenterology. 2002;37(5):345–53. doi: 10.1007/s005350200048. [DOI] [PubMed] [Google Scholar]
- 6.Leon AJ, Gomez E, Garrote JA, et al. High levels of proinflammatory cytokines, but not markers of tissue injury, in unaffected intestinal areas from patients with IBD. Mediators of inflammation. 2009;2009:580450. doi: 10.1155/2009/580450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukada Y, Nakamura T, Iimura M, Iizuka BE, Hayashi N. Cytokine profile in colonic mucosa of ulcerative colitis correlates with disease activity and response to granulocytapheresis. The American journal of gastroenterology. 2002;97(11):2820–8. doi: 10.1111/j.1572-0241.2002.07029.x. [DOI] [PubMed] [Google Scholar]
- 8.Raddatz D, Bockemuhl M, Ramadori G. Quantitative measurement of cytokine mRNA in inflammatory bowel disease: relation to clinical and endoscopic activity and outcome. European journal of gastroenterology & hepatology. 2005;17(5):547–57. doi: 10.1097/00042737-200505000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Matsuda R, Koide T, Tokoro C, et al. Quantitive cytokine mRNA expression profiles in the colonic mucosa of patients with steroid naive ulcerative colitis during active and quiescent disease. Inflammatory bowel diseases. 2009;15(3):328–34. doi: 10.1002/ibd.20759. [DOI] [PubMed] [Google Scholar]
- 10.Arijs I, De Hertogh G, Machiels K, et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. The American journal of gastroenterology. 2011;106(4):748–61. doi: 10.1038/ajg.2011.27. [DOI] [PubMed] [Google Scholar]
- 11.Roman J, Planell N, Lozano JJ, et al. Evaluation of responsive gene expression as a sensitive and specific biomarker in patients with ulcerative colitis. Inflammatory bowel diseases. 2013;19(2):221–9. doi: 10.1002/ibd.23020. [DOI] [PubMed] [Google Scholar]
- 12.Noble CL, Abbas AR, Cornelius J, et al. Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut. 2008;57(10):1398–405. doi: 10.1136/gut.2008.148395. [DOI] [PubMed] [Google Scholar]
- 13.Boyle DL, Rosengren S, Bugbee W, Kavanaugh A, Firestein GS. Quantitative biomarker analysis of synovial gene expression by real-time PCR. Arthritis research & therapy. 2003;5(6):R352–60. doi: 10.1186/ar1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317(26):1625–9. doi: 10.1056/NEJM198712243172603. [DOI] [PubMed] [Google Scholar]
- 15.Reimund JM, Wittersheim C, Dumont S, et al. Mucosal inflammatory cytokine production by intestinal biopsies in patients with ulcerative colitis and Crohn’s disease. Journal of clinical immunology. 1996;16(3):144–50. doi: 10.1007/BF01540912. [DOI] [PubMed] [Google Scholar]
- 16.te Velde AA, de Kort F, Sterrenburg E, et al. Comparative analysis of colonic gene expression of three experimental colitis models mimicking inflammatory bowel disease. Inflammatory bowel diseases. 2007;13(3):325–30. doi: 10.1002/ibd.20079. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi A, Watanabe K, Narumi S, et al. The production of interferon-gamma-inducible protein 10 by granulocytes and monocytes is associated with ulcerative colitis disease activity. Journal of gastroenterology. 2007;42(12):947–56. doi: 10.1007/s00535-007-2118-9. [DOI] [PubMed] [Google Scholar]
- 18.Carlsen HS, Baekkevold ES, Johansen FE, Haraldsen G, Brandtzaeg P. B cell attracting chemokine 1 (CXCL13) and its receptor CXCR5 are expressed in normal and aberrant gut associated lymphoid tissue. Gut. 2002;51(3):364–71. doi: 10.1136/gut.51.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis E, Ribbens C, Godon A, et al. Increased production of matrix metalloproteinase-3 and tissue inhibitor of metalloproteinase-1 by inflamed mucosa in inflammatory bowel disease. Clinical and experimental immunology. 2000;120(2):241–6. doi: 10.1046/j.1365-2249.2000.01227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth J, Goebeler M, Sorg C. S100A8 and S100A9 in inflammatory diseases. Lancet. 2001;357(9261):1041. doi: 10.1016/S0140-6736(05)71610-X. [DOI] [PubMed] [Google Scholar]
- 21.Kenward MG, Roger JH. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53(3):983–97. [PubMed] [Google Scholar]
- 22.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2013. http://www.R-project.org. [Google Scholar]
- 23.Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.0–4. 2013 http://CRNA.R-project.org/package=lme4.
- 24.Halekoh U, Hojsgaard S. pbkrtest: Parametric bootstrap and Kenward Roger based methods for mixed model comparison. R package version 0.3–7. 2013 http://CRAN.R-project.org/package=pbkrtest.
- 25.Gomes P, du Boulay C, Smith CL, Holdstock G. Relationship between disease activity indices and colonoscopic findings in patients with colonic inflammatory bowel disease. Gut. 1986;27(1):92–5. doi: 10.1136/gut.27.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feagan BG, Sandborn WJ, D’Haens G, et al. The role of centralized reading of endoscopy in a randomized controlled trial of mesalamine for ulcerative colitis. Gastroenterology. 2013;145(1):149–157. e2. doi: 10.1053/j.gastro.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 27.U.S. Department of Health and Human Services FDA, Center for Drug Evaluation and Research. [Accessed 5/22/14];Guidance for Industry: Bioanalytical Method Validation. 2013 Sep; Available at: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM368107.pdf.
- 28.Olsen T, Goll R, Cui G, Christiansen I, Florholmen J. TNF-alpha gene expression in colorectal mucosa as a predictor of remission after induction therapy with infliximab in ulcerative colitis. Cytokine. 2009;46(2):222–7. doi: 10.1016/j.cyto.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Buch MH, Boyle DL, Rosengren S, et al. Mode of action of abatacept in rheumatoid arthritis patients having failed tumour necrosis factor blockade: a histological, gene expression and dynamic magnetic resonance imaging pilot study. Annals of the rheumatic diseases. 2009;68(7):1220–7. doi: 10.1136/ard.2008.091876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kavanaugh A, Rosengren S, Lee SJ, et al. Assessment of rituximab’s immunomodulatory synovial effects (ARISE trial). 1: clinical and synovial biomarker results. Annals of the rheumatic diseases. 2008;67(3):402–8. doi: 10.1136/ard.2007.074229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.