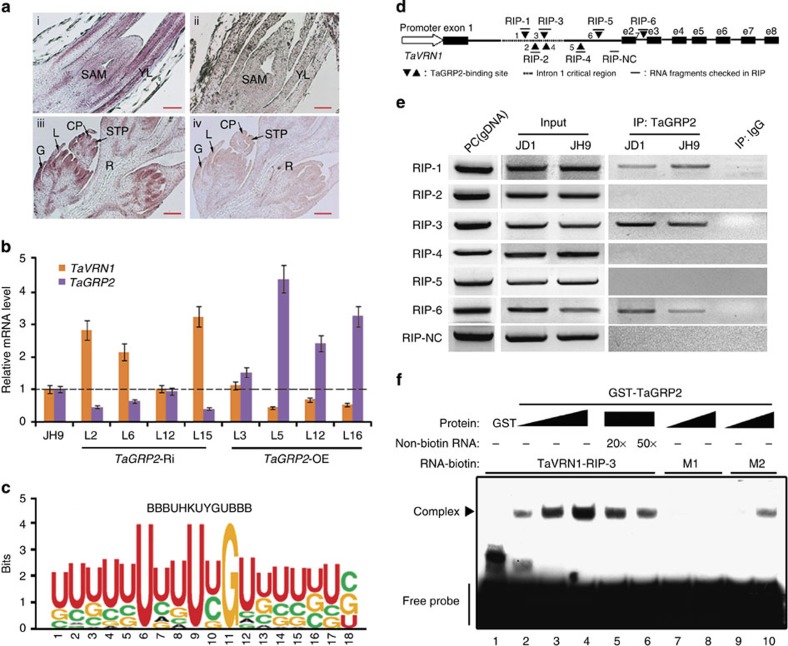
Figure 5. TaGRP2 represses TaVRN1 expression by directly binding its pre-mRNA.
(a) Expression pattern of TaGRP2 in shoot tips (top) and flower organs (bottom). RNA in situ hybridization using aTaGRP2 antisense (left) or sense probe (right). SAM, shoot apical meristem; YL, young leaf; G, glume; L, lemma; CP, carpel primordium; STP, stamen primordium; R, rachilla. (b) Transcript accumulation of TaVRN1 and TaGRP2 in TaGRP2-OE, TaGRP2-Ri and JH9 plants. The expression levels of TaVRN1 and TaGRP2 in wild-type JH9 are set as 1. Relative fold change in the transgenic plants. Data are means±s.d. of three parallel samples. (c) Weblogo of the RNA motif bound by TaGRP2 from RNA library sequences obtained by SELEX. (d) Diagram of the putative TaGRP2-binding sites in TaVRN1. Black triangles indicate the positions of putative TaGRP2-binding sites; dashes show the fragment amplified by RT-PCR after RIP using antibody against TaGRP2. e2–e8, exon 2–exon 8; RIP-NC, RIP negative control fragment. (e,f) Demonstration of direct binding of TaGRP2 to TaVRN1 pre-mRNA with conserved binding sites through RIP (e) or RNA-EMSA (f). PC, genomic DNA serves as a positive control. M1 represents RNA probe mutated in the core nucleotide U, U, G; M2 indicates mutation in the adjacent nucleotides.