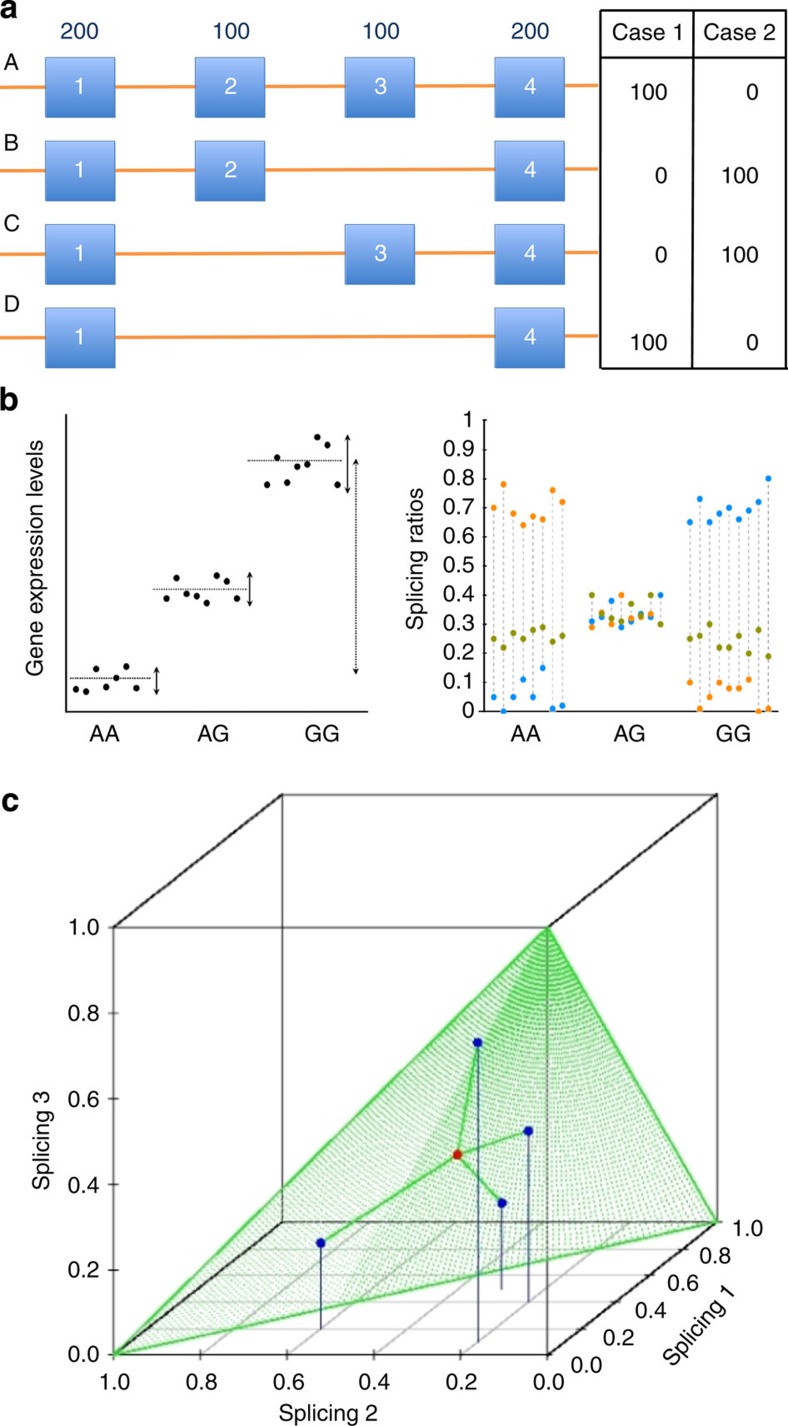
Figure 1. sQTLs and the sQTLseekeR approach.
(a) Alternative transcript versus alternative exon usage. A gene with four isoforms (A,B,C and D), whose abundances is measured in two different individuals. In individual 1, only isoforms A and D are found with abundance 100 (say copies per cell) each. In individual 2, only isoforms B and C are found, also with abundance 100 each. The splicing pattern in the two individuals is completely different. However, exons abundances (on top of the exon, in the figure) are identical in the two individuals. (b) Testing the association between the distribution of the relative abundances of a gene’s transcript isoforms and a SNP. Left panel. The expression level of gene (y axis) measured on individuals polymorphic at a given genomic site (SNP, x axis). There is a clear association between genotype and expression. To test for association between the SNP and gene expression, the variance of the expression within the genotypes (solid arrows) and between the genotypes (dashed arrow) can be compared. Right panel. The abundances of each individual transcript isoform (blue, green and orange) for a gene measured in individuals polymorphic at a genomic site (x axis). The relative abundances of each transcript (splicing ratios) are computed relative to the total abundance of the gene (y axis). There is a clear association between genotypes and splicing ratios. The AA genotypes express predominantly the orange isoform, while the GG genotypes express predominantly the blue isoform. The heterozygous express the two isoforms at similar levels, and similar to the levels of the green isoform—whose relative abundance does not appear to be affected by the genotype. In our approach, association between SNP and splicing ratios is also tested by comparing within and between genotype variabilities of the splicing ratios. See Methods. (c) The space of the splicing ratios of a gene with three isoforms. Four samples/observations (blue points) are represented. The point in red is the centroid of the samples and the green triangle represents the 2-simplex space. The sum of squared distances (SS) between the observations and the centroid is the basic measure of variability used in our approach.