Fig. 3.
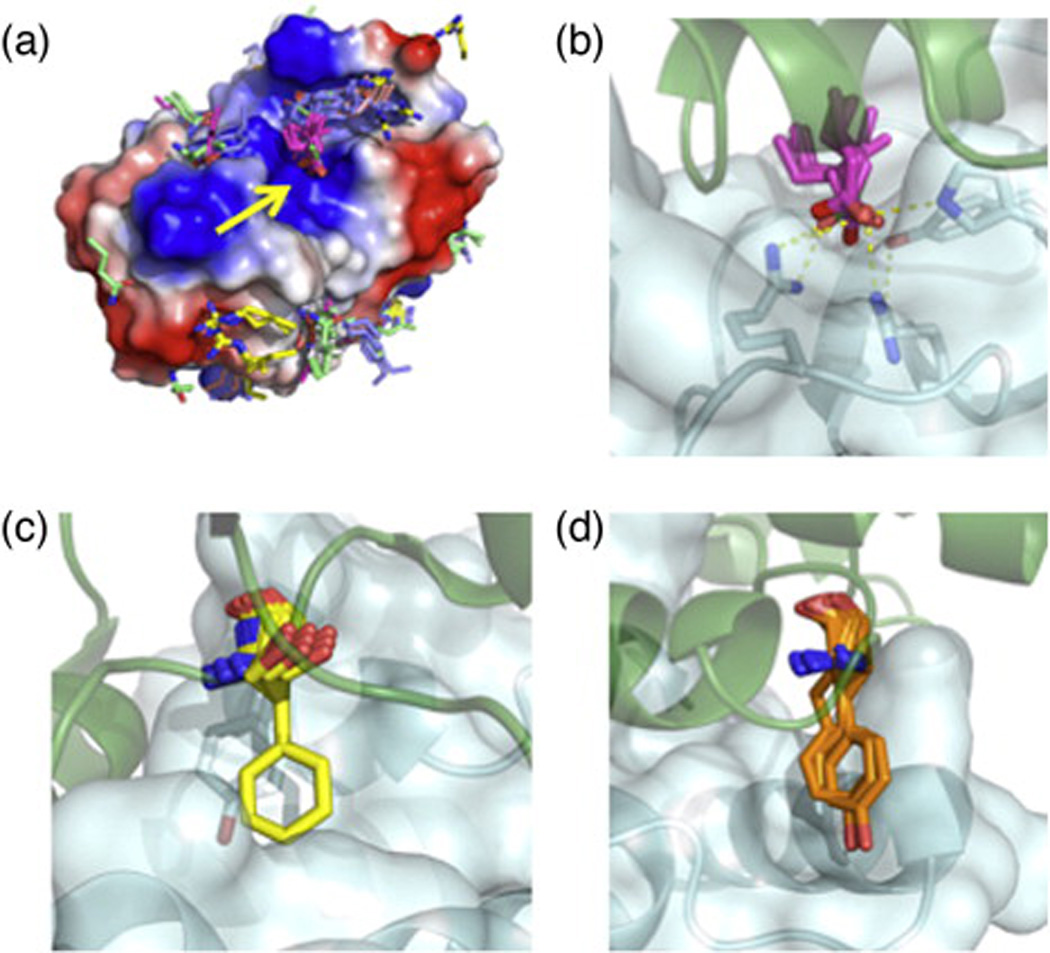
Construction of hotspot residue libraries. (a) A de novo hotspot library of the barnase surface highlights the barstar hotspot-binding site (yellow arrow). Ten thousand trajectories of all natural amino acids except Gly, Cys, and Pro were evaluated, and the best 1% by calculated binding energy of each residue identity was used to build a hotspot library. The barnase surface is colored by electrostatic charge using vacuum electrostatics in PyMOL. (b) De novo identified hotspot interactions of Glu (pink sticks) closely recapitulate the interaction of barstar Asp39 (green sticks) with barnase. (c) Inverse rotamers of the colicin E9 hotspot Phe86 (yellow sticks) against the surface of Im9. Im9 hotspot residue Tyr54 is shown in pale-cyan sticks. Highly optimal π-stacking interactions between E9 Phe86 and Im9 Tyr54 restrict the rigid-body orientation of the Phe hotspot residue. (d) E9 hotspot Tyr83 has more conformational freedom against the Im9 surface than the Phe86 of (c), allowing for rigid-body docking of the Tyr residue (orange). For each docked conformation, inverse rotamers were built as in (c) but, for clarity, are not shown in this figure.
