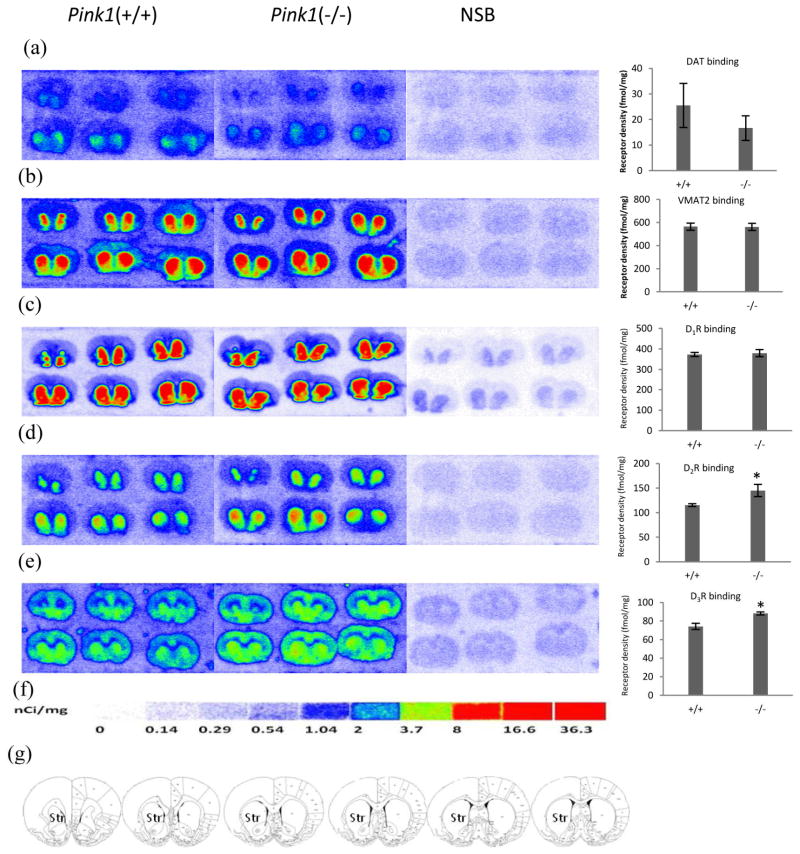
Figure 2. Quantitative autoradiographic analysis of DAT, DTBZ and dopamine receptors densities in the PINK1 gene knockout rat models of PD.
Autoradiograms show binding of 8.5 nM [3H]WIN35428 (a), 23.5 nM [3H]DTBZ (b), 3.8 nM [3H]SCH23390 (c), 4.4 nM [3H]raclopride(d) and 5.9 nM [3H]WC-10 (e) on multiple brain sections through the striatum in the PINK1 gene knockout and wild-type littermate rat. Nonspecific binding was determined in presence of 1 μM nomifensine (for [3H]WIN35428), 1 μM S(−)-tetrabenazine (for [3H]DTBZ),1 μM (+) butaclamol (for [3H]SCH23390), 1 μM S(−)-eticlopride (for [3H]raclopride and [3H]WC-10). DAT, VMAT2 and D1 receptor binding did not change in PINK1 gene knockout rat (a,b,c). The densities of dopamine D2 and D3 receptors were significant upregulated in PINK1 gene knockout rats (d, e). [3H]Microscale standards (ranging from 0 to 36.3 nCi/mg) were also counted (f). Schematic rat brain sections showing the rostral to caudal extent of the striatum, the region of interest in which DAT, VMAT2, D1, D2, and D3 receptors were quantified, across a total of six sections (g). NSB, nonspecific binding; Str, striatum. *p<0.05 for Pink1 knockout rat vs. control rat.