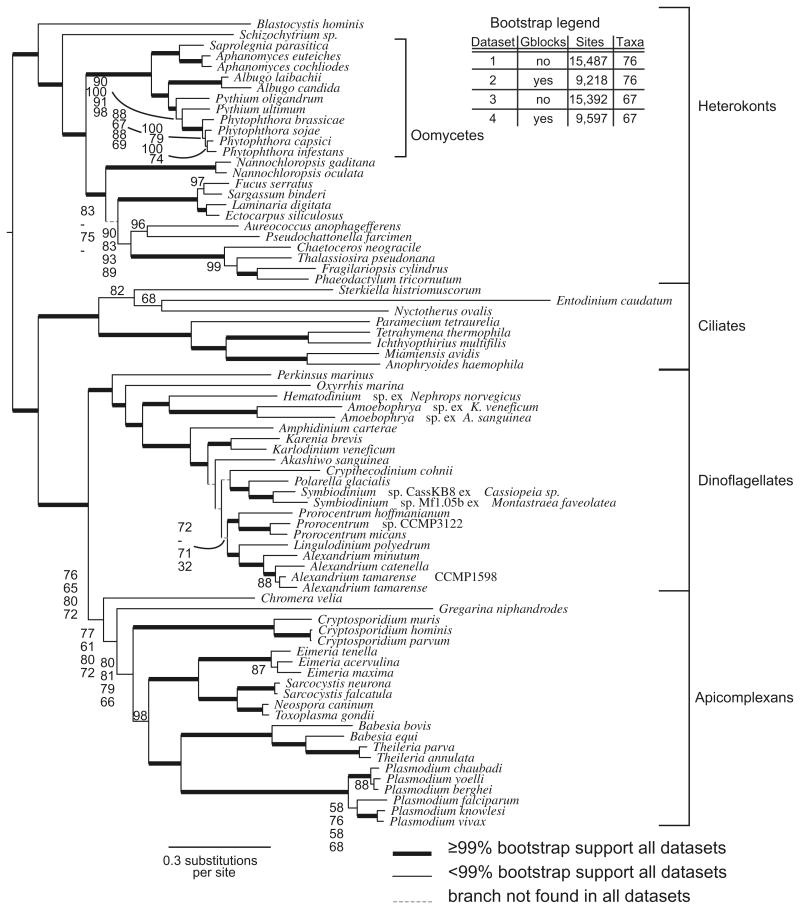
Fig. 2.
The most likely tree found by RAxML using the LG amino acid substitution matrix with gamma correction and 500 rapid bootstraps. Bold branches were found in 100% of bootstrap replicates. The dataset was partitioned both by using gblocks and by removing the 9 taxa with fewer aligned characters as shown on the legend in the figure. For branches with variable support in different partitions of the alignment (>5% difference in bootstrap values) the values are shown for all four different alignments.