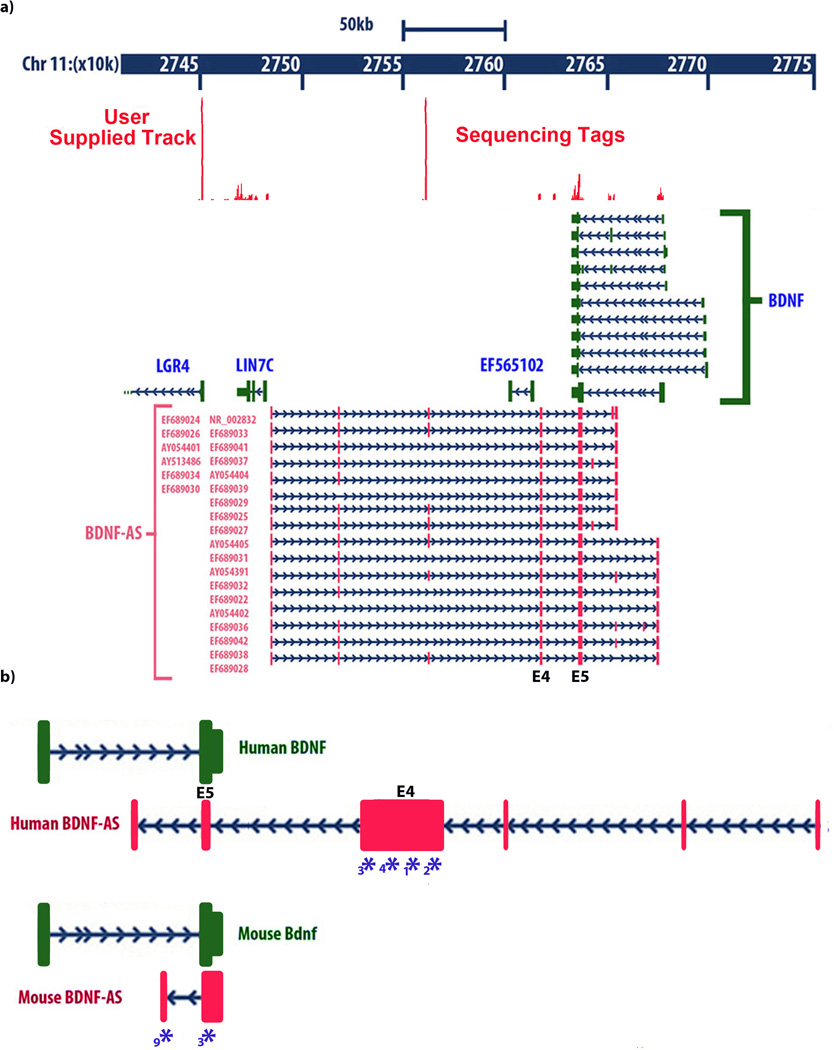
Figure 1. Genomic organization of the human BDNF locus showing.
(A) genomic location of the BDNF sense and antisense transcripts and their relation to the other neighboring genes on chromosome 11. Solid boxes show exons and arrows show introns and direction of transcription. Different splice variants of BDNF-AS transcript are transcribed from the opposite DNA strand as that of BDNF mRNA. All BDNF-AS splice variants have a common exon that overlaps with 225 bp of all variants of BDNF mRNA. Inset data: Sequence tags generated by next-generation sequencing (RNA deep-seq), derived from human entorhinal cortex are aligned to the UCSC genome browser. Peaks represent nucleotide coverage, indicating reliable detection of BDNF-AS exons.
(B) Identification of mouse Bdnf-AS by RACE experiments followed by sequencing. Proportional drawing representing human and mouse BDNF loci, showing direction of transcription for both BDNF and BDNF-AS transcripts, as well as the potential overlapping region. Mouse Bdnf-AS transcript is shorter than that of human and contains 1–2 exons. The potential overlapping region is bigger in mouse (934 bp compared to 225 bp in human), but the common overlap shows 90% homology across the two species. Real-time PCR (RT-PCR) primers, probes, and siRNAs are designed to target the non-overlapping parts of both transcripts. Blue Asterisks and numbers below each denote target sites of siRNAs used in this study, as well as mBdnf-AntagoNAT3(*3) and mBdnf-AntagoNAT9 (*9).