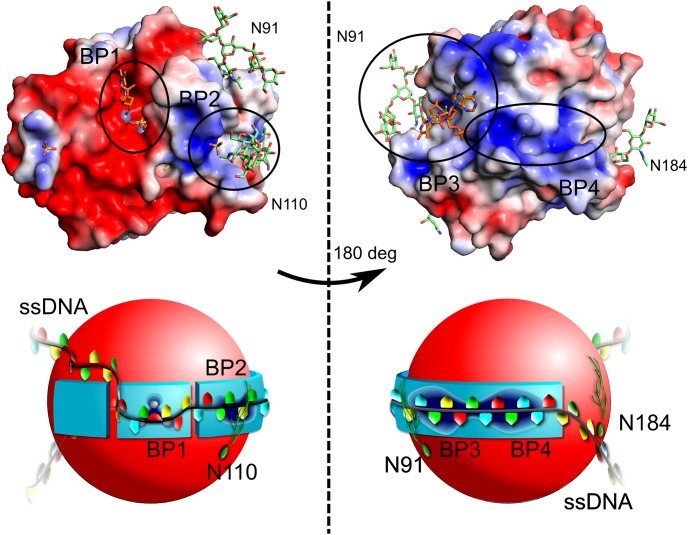
Figure 5. The ssDNA binding pockets and wrapped ssDNA binding model.
Top: DelPhi electrostatic surface ranging from -3 (deep red) to 3 (deep blue). Zn is colored gray, and was not used in the electrostatic potential calculations. The model was generated by combining the well-defined glycan structures of 4CXP (green) with the ssDNA ligands of 4CXO (orange). The protein is presented from the active site side, on the left, and rotated by 180 degrees, on the right. Bottom: Schematic representation of the above model. In total, we identified two experimental binding pockets (BP1 and BP3), and we propose two more (BP2 and BP4). BP1 is the active site, in which the ssDNA hydrolysis takes place. BP2 is lined by Trp43, Arg46, Arg50, and GlcNAc1 from N-glycan at position 110. BP3 is the large secondary ssDNA binding site, which was determined in our crystal structure. It is described in greater detail in Fig. 4 and is well conserved in our alignment. BP4 is a positive charge-rich extension of BP3, which in addition contains three tryptophans and one tyrosine (Fig. S2). We propose that the ssDNA will wrap itself around the AtBFN2 molecule via BP2, BP3, and BP4, with further stabilization provided by the N-glycans (green filaments in our figure). The reaction will then take place in BP1.