Table 1. Characterization of the QTL regions.
| SSC1 | Position (Mb) | Most sign. SNP | MAF | MGF | −log10 (Pvalue) | SNP effects2 | SNP variance (% of 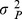 ) ) |
||||
| Start | End |
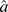
|
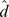
|
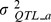
|
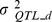
|
||||||
| 4 | 44.53 | - | 44.53 | ASGA0019540 | 0.31 | 0.09 | 4.27 | 0.04 | −0.26* | 0.13 | 1.14 |
| 6 | 101.77 | - | 104.41 | ALGA0036369 | 0.36 | 0.14 | 6.37 | 0.27* | −0.18 | 1.97 | 0.60 |
| 7 | 103.03 | - | 103.59 | ASGA0035500 | 0.32 | 0.12 | 7.59 | 0.29* | 0.04 | 3.02 | 0.02 |
| 12 | 52.71 | - | 54.68 | ALGA0120076 | 0.43 | 0.20 | 5.19 | 0.24* | 0.05 | 2.25 | 0.06 |
| Total | 7.37 | 1.82 | |||||||||
Minor allele frequency (MAF), minor genotypic frequency (MGF), −log10 (P-values) of the association analysis, additive (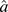 ) and dominance (
) and dominance (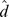 ) effect estimates, proportion of the total phenotypic variance (
) effect estimates, proportion of the total phenotypic variance (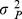 ) explained by the additive and dominance variances (
) explained by the additive and dominance variances (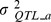 and
and 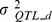 ) based on the most significant SNP of each QTL region.
) based on the most significant SNP of each QTL region.
SSC: Sus scrofa chromosome;
*Contrast to evaluate the mode of gene action was significant (P<0.01);
Additive effects expressed in absolute values.
