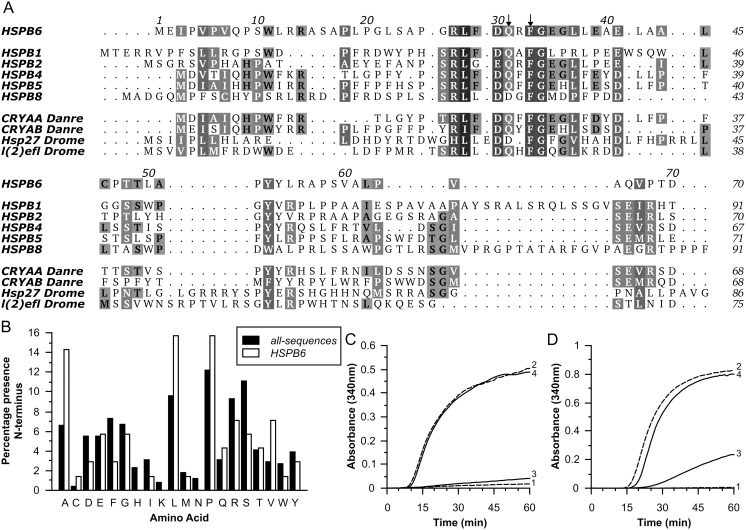
Figure 1. Properties of the N-terminal domain of HSPB6.
(A) Multiple sequence alignment of the NTD of HSPB6 with other human sHSPs, the α-crystallins from Danio rerio (CRYAA and CRYAB) and two related proteins from Drosophila melanogaster (l(2)_efl and Hsp27). The two highly conserved residues that are mutated in this work are indicated by arrowheads. The alignment was made using Aline [54]. Uniprot accession numbers: HSPB1 (Hsp27): P04792, HSPB2: Q16082, HSPB4 (CRYAA): P02489, HSPB5 (CRYAB): P02511, HSPB6: O14558, HSPB8: Q9UJY1, CRYAA_Danre: Q8UUZ6, CRYAB_Danre: Q9PUR2, Hsp27_Drome: P02518, l(2)efl_Drome: P82147. (B) Amino acid composition within the N-terminus of HSPB1, HSPB2, HSPB4, HSPB5, HSPB6 and HSPB8 and orthologues in different species (H. sapiens, B. taurus, M. musculus, X. leavis or tropicalis, G. gallus and D. rerio). The average amino acid composition is shown as white bars, HSPB6 amino acid composition is shown as black bars. (C–D) Chaperone-like activity of human HSPB6 and the N-terminal deletion construct (ΔN) using insulin (C) and yADH (D) as substrates. The monomer mass molar ratios of substrate:sHSP were 1∶0.2 and 1∶2 respectively, and were the same as used in further experiments. Curve 1, substrate alone; 2, substrate + DTT; 3, with addition of HSPB6; 4, with addition of ΔN.