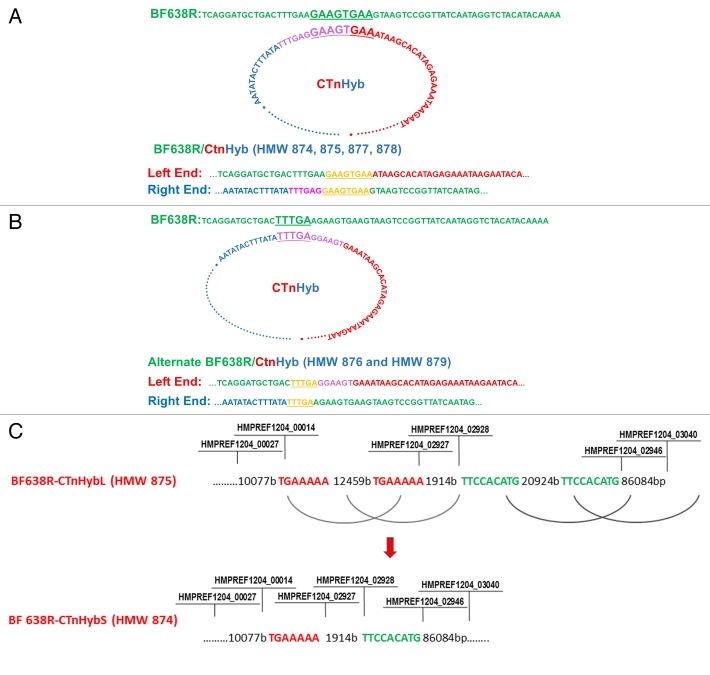
Figure 3. (A) Predicted events leading to integration of CTnHyb into the BF638R chromosome to result in BF638R/CTnHyb. The circular form of CTnHyb recombines with BF638R at the underlined sequence (GAAAGTGAA). (B) Alternate integration of CTnHyb into the BF638R chromosome. Predicted events leading to integration of CTnHyb into the BF638R chromosome to result in BF638R/CTnHyb found in HMW 876 and HMW 879 . The circular form of CTnHyb recombines with BF638R at the underlined sequence (TTTGA). (C) Predicted model of the deletions in CTnHybL leading to CTnHybS. The locus tag labels serve as approximate reference points. The bases are referred to as “bases” or “b”. The regions of homology that are predicted to recombine are color coded. The nimJ gene is among the genes deleted in the first deletion and metronidazole has a lower MIC for BF638R/CTnHybS than for BF638R/CTnHybL, as expected (Table 1).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.