Abstract
It is increasingly recognized that infiltrating immune cells contribute to the pathogenesis of a wide range of solid tumors. The paracrine signaling between the tumor and the immune cells alters the functional state of individual tumor cells and, correspondingly, the anticipated response to radiation or chemotherapies, which is of great importance to clinical oncology. Here we present a high-density microchip platform capable of measuring a panel of paracrine signals associated with heterotypic tumor-immune cell interactions in the single-cell, pair-wise manner. The device features a high-content cell capture array of 5000+ sub-nanoliter microchambers for the isolation of single and multi- cell combinations and a multi-plex antibody “barcode” array for multiplexed protein secretion analysis from each microchamber. In this work, we measured a panel of 16 proteins produced from individual glioma cells, individual macrophage cells and varying heterotypic multi-cell combinations of both on the same device. The results show changes of tumor cell functional phenotypes that cannot be explained by an additive effect from isolated single cells and, presumably, can be attributed to the paracrine signaling between macrophage and glioma cells. The protein correlation analysis reveals the key signaling nodes altered by tumor-macrophage communication. This platform enables the novel pair-wise interrogation of heterotypic cell-cell paracrine signaling at the individual cell level with an in-depth analysis of the changing functional phenotypes for different co-culture cell combinations.
Introduction
A solid tumor is comprised of not only tumor cells but also stromal and infiltrating immune cells.1,2 The intercellular signaling network established between these diverse cell types collectively shapes a complex tumor microenvironment and can alter tumor progression or therapeutic response over time.3–12 Approaches that can interrogate multiple cell types as well as examine the cell-cell communication network mediated by an array of soluble paracrine signaling molecules, e.g., cytokines, growth factors, and neuropoientins9,13–16 will improve our understanding of disease mechanism and potentially lead to the development of new therapeutic strategies by targeting the complex microenvironments.13,17,18 Prior to moving into the modeling of complex tumor microenvironment, a fundamental question is how to quantify tumor-immune paracrine communication in the single-cell pair-wise manner and at the systems level.
Recently, microchip platforms have been developed for controlled assembly of heterotypic cell pairs. Qin et al. reported on a block-cell-printing method to assemble different tumor cells and neurons in a highly controlled, pair-wise manner.19 Voldman et al. used a microfluidic hydrodynamic trapping microchip to create pairs of mouse embryonic fibroblast and stem cells and further induced their fusion on chip.20 Although cell-cell interactions such as filopodia junction and cell fusion have been demonstrated, it remains challenging to measure all paracrine signals, which are secreted factors, in these individual heterotypic cell pairs. On the other hand, exemplary “lab-on-a-chip” platforms have been developed for quantitative analysis of protein secretion from single immune and cancer cells.21–22 Love et al. developed microengraving methodology to quantify secretion for up to four cytokines from single viable primary immune cells.23 We previously demonstrated a microchip platform capable of measuring up to 15 cytokines from single tumor cells on chip.22 One of the recent approaches developed by Heath et al. utilized a microchip to investigate growth factor-driven protein signaling dependence on the distance between the same type of cancer cells.24,25 While each of these systems and alternative co-culture methodologies attempt to measure either autocrine proteins from individual cells or a limited number of paracrine factors from homotypic pairs of tumor cells, the study of a large array of heterotypic cell pairs and their paracrine signals has not been reported.
Herein, we present a microchip platform, which was built upon our previous high-throughput single cell secretomic microchip.22 We demonstrate the measurement of 16 secreted proteins in a large array of subnanoliter microchambers containing individual glioma cells, individual macrophage cells, or varying combinations of both on the same device. This simple device, which has 5000+ microchambers, does not require precise control of cell trapping, but allows for creating hundreds of individual tumor-macrophage pairs simply through a random-loading method. The results revealed distinct functional heterogeneity among glioma cells, which is altered significantly by the addition of individual macrophages in the same microchamber, which can not be qualitatively interpreted as the additive effect and indicates resolvable paracrine signaling interactions. The key protein clusters can be identified by a protein correlation analysis.
Results
Population level analysis of cell-cell paracrine between glioma cells and macrophages
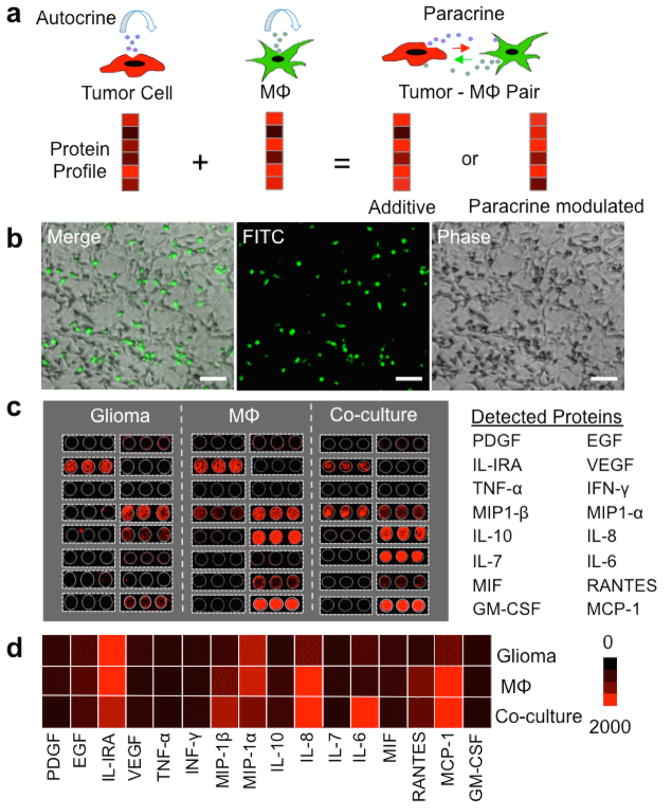
As a first attempt to assess cell-cell paracrine in the context of tumor microenvironments, we focus on the interaction between glioma cells and macrophage, a phenotypic equivalent of microglia in the central nervous system. In order to outline the major functional proteins associated with glioma-macrophage interaction and establish the anticipated outcome at the population level, we start with a bulk assay of protein secretion from human glioma cells, macrophages and their co-cultures. U-87 glioma cells, U937-differentiated macrophages and their co-cultures were loaded in a microwell-based cell culture platform developed in-house (Figure 1b, Supporting Figure 1). In each culture device, there were 3 large (D~ 1 cm) microwell partitions for “glioma alone”, “macrophage alone” and “glioma-macrophage interaction” experiments side-by-side (Figure 1b, Supporting Figure 1). The functions and full names of these proteins are summarized in Supporting Tables 1. Glioma cells, macrophages and their mixture were grown, respectively, in the culture and co-culture microwells, imaged with motorized fluorescence microscope, up to 24 hours. In the co-culture microwell, prior to the experiment, macrophage cells were stained with green live cell tracker dyes (CMFDA, Invitrogen). The supernatant was collected from these cell cultures after 24 hours for measuring 16 secreted proteins using a conventional pin-spotted microarray (Supporting Figure 2). From each cell culture well, 50 μL of sample was collected to measure population secretion profile, and 100-μL of fresh medium was added back into each well to maintain the cultures. The representative results of antibody microarray analysis of protein secretion for 24 hours from all three cell cultures – glioma, macrophage and glioma-macrophage co-culture – are shown in Figure 1c (also Supporting Figure 2). Each protein was measured in triplicate by three antibody spots for each culture and the average fluorescence intensities are shown as a heatmap in Figure 1d. Glioma cells secreted chemoattractant (MIP-1α, MCP-1), pro-inflammatory (IL-1RA, IL-6), pro-angiogenesis (IL-8), and proliferation (EGF) factors. Macrophages, on average, also secreted IL-IRA, MIP-1α, IL-8, and MCP-1, but differentially expressed MIP-1β and RANTES. The protein secretion pattern of glioma-macrophage co-culture was related to individual glioma or macrophage population secretion, but not a simple additive response. The secretion levels of IL1RA and MIP-1α were reduced in co-culture. IL-6 and MIP-1β were markedly increased in the co-cultured system. The results indicate the involvement of paracrine signaling between the two cell types.
Figure 1. Population level assay of glioma-macrophage paracrine signaling.
(a) Schematic depiction of tumor-macrophage paracrine signaling changing protein profiles at the single cell level. (b) Optical images showing co-culture of human glioma cells (U87) and macrophages (U937-derived) in a conventional microwell platform. Macrophages were pre-stained with green cell tracker dye CMFDA and visualized using the FITC fluorescence channel. Scale bar: 100 μm. (c) Scanned fluorescent images of the pin-spotted antibody microarrays used to measure protein secretion. Cell culture media were collected after 24 hours of incubation of glioma, macrophage (Mϕ) and glioma-macrophage co-culture. Microarrays were read out using APC dye-labeled streptavidin at 635 nm (Red). (d) Heat map showing 24-hour protein secretion profiles of glioma, macrophage (Mϕ) and glioma-macrophage cultures at population level.
Single-cell level analysis of cell-cell paracrine between glioma cells and macrophages
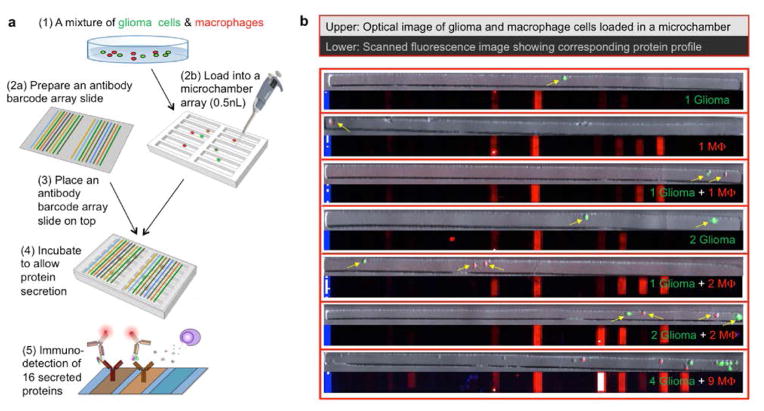
Due to intratumor cellular heterogeneity and non-genetic cell-cell variability, the functional states of individual tumor cells and tumor-associated immune cells isolated from the same tumor tissue can vary substantially. Therefore, to accurately quantify the paracrine signals (secreted proteins) and their role in modulating cellular response, there is a need for conducting such study at the individual cell level.26–28 Our previously reported single-cell secretomic analysis microchip platform22 was modified to measure protein secretion profiles of single glioma cells, single macrophages and varying multi-cell combinations of both cell types on the same chip in different isolated microchambers (Figure 2). Briefly, this microchip consists of two separate components: (1) a high-density subnanoliter microchamber array fabricated from polydimethylsiloxane (PDMS) to capture and isolate single cells or low count multi-cell combinations over a multiplexed antibody array, and (2) a high-density antibody barcode array patterned on a poly-L-lysine functionalized glass slide via a custom flow patterning technique.22, 29 The total number of microchambers is ~5500 with each chamber measuring 35 μm x 35 μm x 1850 μm in length x depth x width. Prior to using this microchip for single-cell level protein assay, we performed rigorous validation experiments to establish the analytic metrics and technological validity. First, we conducted cross-reactivity check using recombinant proteins to ensure the antibody pairs are specific in this multiplex immunoassay format (Supporting Figure 3). Second, we further performed titration experiments (Supporting Figure 4) to valid the antibodies for the limit of detection and dynamic range. All antibodies used in the study and their source clones are summarized in Supporting Table 2.
Figure 2. Microchip for protein secretion profiling of glioma and macrophage cells at the single-cell level.
(a) Work flow of high-throughput multiplexed single cell secretomic assay to measure glioma, macrophage and their combinations. (b) Optical micrograph of single macrophage (red), glioma (green) and glioma- macrophage co-culture microwells and their corresponding scanned fluorescence images showing the raw data of single cell secretomic measurements. Blue lines are position marks created with immobilized Fluorescein Isothiocyanate labeled bovine serum albumin (FITC–BSA, 488). Red is APC dye-streptavidin (Cy5, 635) for specific protein detection within each microwell.
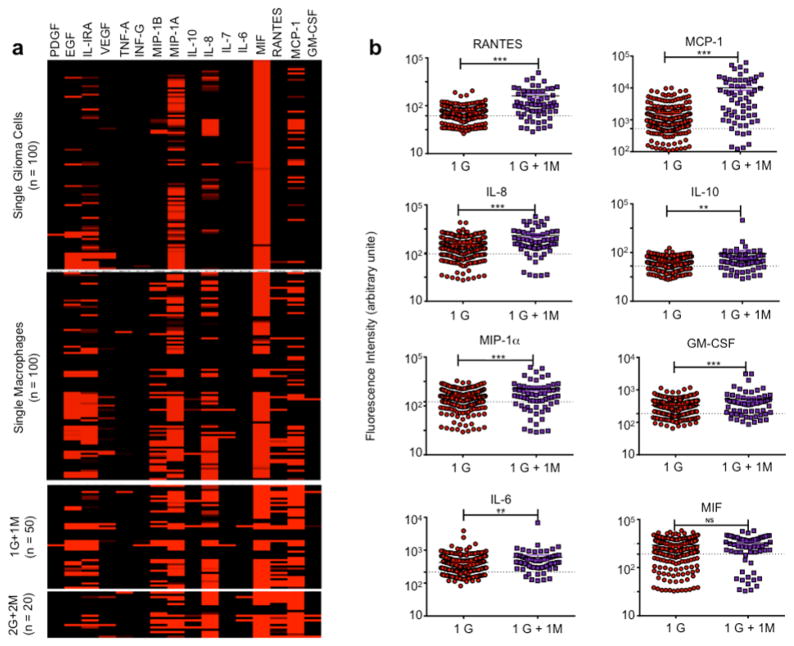
In this study, the microchamber component is loaded with a mixture of glioma and macrophage cells and then the loaded array is subsequently sandwiched with an antibody barcode slide such that each individual is placed in a conventional tissue incubator for ~20 hours to allow for cell secretion. Afterwards, the antibody barcode glass slide was removed and the barcode array was developed by introducing a cocktail of detection antibodies and then fluorescent probes for subsequent protein detection. Representative images of 7 microchambers with cells loaded and the corresponding protein secretion files detected by the fluorescent barcode images are shown in Figure 2b. Analysis of fluorescence intensity of the antibody barcode array corresponding to the 16 proteins per microchamber resulted in a quantitative data set showing protein secretion profiles associated with single glioma cells, single macrophages and their varying multi-cell combinations. Figure 3a is a heat map of the secreted proteins from microchambers with single glioma cells, with single macrophages, with single-cell pairs (1 glioma + 1 macrophage) and two-cell pairs (2 glioma + 2 macrophage). Among single glioma cells, expression of MIF or MIP-1α protein was high. IL-8, EGF, IL-1RA, and MCP-1 were present. Single macrophage cells, in general, produced more proteins. “1 glioma + 1 macrophage” microchambers produced higher levels of MCP-1 and RANTES. When the cell number increased to 2 for both types of cells, the protein profile seeming did not change significantly except emergence of IL-6 secretion (Supporting Figures 6–8). However, the presence of a glioma cell with a macrophage instead of glioma cells alone in a microwell significantly changed the expression level of RANTES, MCP-1, MIP-1α, GM-CSF, IL-8 (*** p < 0.001), IL-10 and IL-6 (** p < 0.01) (Figure 3b, Supporting Figure 5), which agreed in part with the population data (Figure 1d). Due to the large variation of single cell protein secretion, more rigorous computational analyses need to be performed to examine the paracrine-induced protein signature alteration at the individual cell level.
Figure 3. Protein secretion levels measured in single glioma cell, single macrophage cell and their combinations.
(a) Heat map showing the profile of 16 proteins secreted from 1 glioma cell (n =100), 1 macrophage cell (n = 100), 1 glioma + 1 macrophage cells (n = 50), and 2 glioma + 2 macrophage cells (n = 20), respectively, as counted within isolated microwells in the custom microchip device. Each row represents single cells while each column presents the protein of interest as corresponding to that particular microwell. (b) Scatter plots comparing protein profiles of RANTES, MCP-1, MIP-1A, GM-CSF, IL-8, IL-10, IL-6, MIF for 1 glioma (1G) vs. 1 glioma + 1 macrophage (1 G + 1 M) for 20 hours incubation (* P < 0.05, ** P < 0.01, *** P < 0.001). The dashed line shows the threshold defined as mean of empty microchamber serving as on-chip controls + 2STDV (standard deviation) to gate true cytokine-secreting cells versus non-secreting cells.
Dependence of the individual protein secretion on glioma-macrophage interaction
We quantitatively examined the average protein secretion levels as a function of the number of heterotypic cells in the same microchambers. Comparing cell grouping combinations, i.e. “1 glioma + 1 macrophage,” “2+2” etc., our results indicate that expression profiles of each cytokine were not linearly additive from single cell results (Supporting Figure 6). For instance, expression levels of MCP-1 did not significantly change in “1+1” or “2+2” pairs, and observed opposite trends in “2+1” and “3+1” groupings (Supporting Figure 7, *p<0.05), IL-6 expression is marked increased in the heterotypic cell pairs as compared to homotypic glioma or macrophage cells (** p < 0.01). We attribute this non-additive expression profile to paracrine signaling interactions between glioma and macrophage cells in the microwell environment, demonstrating the utility of this platform to analyze the impact of cell communication on functional phenotypes by using different building blocks of the tumor microenvironment.
Protein correlation analysis
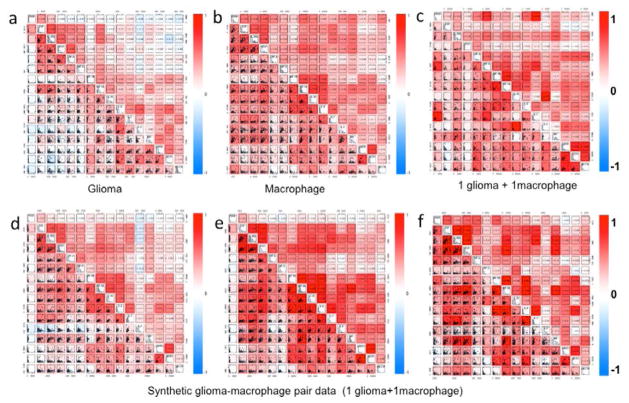
To investigate the global relationship of all expressed proteins at the single-cell level, we performed a pairwise correlation analysis analogous to flow cytometry with linear regression included expressed as a 16x16 cytokine matrix for each cell (Figure 4). The protein correlation matrix is presented to reveal functional clusters by correlated secretions. The protein correlation map for single glioma cells (Figure 4a) exhibits two clusters; growth factors (upper left) and proinflammatory cytokines and chemokines (lower right), which work in concert during glioma response. Single macrophages observe a clustered, but distinct, pattern (Figure 4b). In both populations, a positive MIP-1α/β, IL-8 and IL-10 cluster was observed. The protein correlation matrix for “1 glioma + 1 macrophage” measured in the same microchip is shown in Figure 4c. To address the “1 glioma + 1 macrophage” non-additive secretomic profile and develop a method to examine the role of cell-cell communication in altering cell function, we took “glioma” data and “macrophage” data to generate a synthetic “glioma-macrophage pair’ from randomized, additive signals from each data set and then compared to the true “1 glioma + 1 macrophage” pair from our microchip device. Three representative correlation maps from the synthetic “1 glioma + 1 macrophage” pair data is shown in Figures 4d–f (d: low R- values; e: high R-values; f: median R-values from randomized pairs). All three panels observe similar cluster correlated directly to typical glioma and macrophage signaling clusters. However, when comparing synthetic glioma- macrophage pair data to the experimental “1 glioma + 1 macrophage,” we observed significant difference beyond standard data variation. In the experimental data, the upper-left cluster is diminished and a tight cluster comprised of RANTES, MCP-1 and GM-CSF, indicative of possible glioma cell initiated communication. The results confirm the existence of glioma-macrophage communication at the single cell level, providing a possible starting point for analyzing individual cell-cell network links in regards to functional heterogeneity and cell type contributions in heterotypic cell interactions.
Figure 4. Protein correlation analysis.
Scatter plot matrices demonstrates protein protein correlation in single cells for (a) glioma, (b) macrophage, (c) “1 glioma + 1 macrophage” and three (d–f) “synthetic 1 glioma + 1 macrophage” pair data. The proteins are listed at the diagonal line. Linear regression analysis was performed to obtain correlation coefficient; R. R-value is proportional with the color intensity where red and blue indicated positive and negative correlations, respectively.
Discussions
We have implemented a microchip platform for profiling a panel of 16 paracrine signaling proteins from single glioma cells, single macrophages and multi-cell heterotypic cell combinations. Although the phenotypically stable cell lines were used in this study, non-genetic cell-cell variability is ubiquitous and may contribute to significant variation of cellular functions including protein secretion.30 First, to confirm protein profile changes owing to pair-wise paracrine induced signaling, the type of cells needs to be identified, which was realized by live cell tracking staining in this study but can be achieved using antibodies against cell specific surface antigen (e.g., EpCAM for tumor cells, CD11b for macrophage cells, and CD3 for T lymphocytes). Second, protein detection sensitivity and quantitative data generation needs to be ensured. Since the amount of secreted proteins is at single cell level and correlated with cell state, cellular heterogeneity and device sensitivity plays an essential role. Recent studies underline the importance of single cell approaches especially in the context of single cell functional proteomics.26–28, 31 Although the nature of the problem is highly complex, one of the requirements from these technologies are being simple and user-friendly while providing high content and high-throughput quantitative data. The microchip technology, that we have, provides these features, allows for creating statistically large number of tumor-macrophage cell pairs on chip with no need for sophisticated microfluidic cell trapping, and permits highly multiplexed quantification of paracrine-induced changes of protein secretion signatures through a pair-wise tumor-macrophage communication. The protein correlation analysis obtained between different cell types for different proteins are in agreement with their known functional classifications, revealing previously unexplored protein correlations that may contribute to tumor function.
Currently, this platform uniquely addresses the need in systems biology to investigate the fundamental mechanism of paracrine-induced changes of cellular functions in a heterotypic cell system and at the single cell level. It can be extended to examine the systems containing more than two types of cells as long as the different cells can be differentially labeled with different fluorescence probes. While the ultimate goal is to investigate the cell-cell communication network in complex tumor microenvironment and examine its role in therapeutic response, there are several limitations that need to overcome in the future development. First, the microchambers were loaded with individual cells in culture medium only, which do not recapitulate the three-dimensional (3D) tumor tissue architecture. Incorporating a cell-laden 3D hydrogel in the microchambers 32 will further improve its ability to study more physiologically relevant tumor-macrophage interactions. Second, the current microchip, although simple and reliable, does not have fluidics for combinational delivery of drug for high-throughput screening of cell response. This technology, as of the current form, is more suited to interrogating individual cells and their combinations from ex vivo specimens or in vitro cell signaling studies.
Supplementary Material
Acknowledgments
The authors thank Dr. Yu Wu for his feedback and assistance in preliminary experiments. This study was supported by the NIH LINCS Program Technology Center Grant (NIH Grant U01 CA164252; PI, R.F.), the U.S. National Cancer Institute Howard Temin Pathway to Independence Award (NIH Grant R00 CA136759; PI, R.F.), and the Single Cell Profiling Core supported by the NIH Grant U54 CA143798 (subaward PI, R.F.). We also acknowledge the Yale Institute for Nanoscience and Quantum Engineering (YINQE) and the Yale Nanofabrication Center to allow us to use their facilities.
Footnotes
Author Contributions
ME conducted single cell, population secretion experiments and cross-reactivity test and data analysis. KB conducted cross-reactivity experiments and data analysis. YL conducted titration experiments. JJC conducted computational analysis of protein correlation matrices. RF conceived the project and conducted data analysis. ME, KB and RF wrote the manuscript.
Conflict of Interest
KB and RF are cofounders of Isoplexis Inc, a company with business interest in developing tools to measure single-cell secretomic functions.
Footnotes should appear here. These might include comments relevant to but not central to the matter under discussion, limited experimental and spectral data, and crystallographic data.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Junttila MR, de Sauvage FJ. Nat Review. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro JR, Yung WKA, Shapiro WR. Cancer Res. 1981;41:2349–59. [PubMed] [Google Scholar]
- 4.Wikstrand CJ, Bigner SH, Bigner DD. Cancer Res. 1983;43:3327–34. [PubMed] [Google Scholar]
- 5.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Genes Dev. 2007;21:2683–26710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 7.Iwasa Y, Michor F. PLoS One. 2011;6:e17866. doi: 10.1371/journal.pone.0017866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg RA. Nat Genet. 2008;40:494–495. doi: 10.1038/ng0508-494. [DOI] [PubMed] [Google Scholar]
- 9.Tlsty TD, Coussens LM. Annu Rev Pathol Mech Dis. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K, Haviv I, Campbell IG. Trends Genet. 2008;25:30–38. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Sawyers CL. Nat Med. 2007;13:1144–1145. doi: 10.1038/nm1007-1144. [DOI] [PubMed] [Google Scholar]
- 12.Kravchenko-Balasha N, Wang J, Remacle F, Levine RD, Heath JR. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1404462111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlou M, Tzelepi V, Efstathiou E. Nat Rev Urol. 2010;7:494–509. doi: 10.1038/nrurol.2010.134. [DOI] [PubMed] [Google Scholar]
- 14.Joyce JA. Cancer Cell. 2005;7:513–520. doi: 10.1016/j.ccr.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Hu M, Polyak K. Curr Opin Dev. 2008;18:27–34. doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zervantonakis IK, Kothapalli CR, Chung S, Sudo R, Kamm RD. Bio Microfluidics. 2011;5(1):013406. doi: 10.1063/1.3553237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Lu Y, Chen W, Fu J, Fan R. PLoS Comput Biol. 2012;8:1002344–10. doi: 10.1371/journal.pcbi.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, Olson OC, Quick ML, Huse JT, Teijeiro V, Setty M, Leslie CS, Oei Y, Pedraza A, Zhang J, Brennan CW, Sutton JC, Holland EC, Daniel D, Joyce JA. Nat Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, Chou CK, Xia X, Hung MC, Qin L. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1313661111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Przybyla L, Voldman J. Annu Rev Anal Chem. 2012;5:293–315. doi: 10.1146/annurev-anchem-062011-143122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin A, Ozawa T, Tajiri K, Obata T, Kondo S, Kinoshita K, Kadowaki S, Takahashi K, Sugiyama T, Kishi H, Muraguchi A. Nat Med. 2009;15(9):1088–1092. doi: 10.1038/nm.1966. [DOI] [PubMed] [Google Scholar]
- 22.Lu Y, Chen JJ, Mu L, Xu Q, Wu Y, Wu PH, Li J, Vortmeyer AO, Miller-Jensen K, Wirtz D, Fan R. Anal Chem. 2013;85:2548–2556. doi: 10.1021/ac400082e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab Chip. 2010;10(11):1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu GC, Ribas A, Heath JR. Nat Med. 2011;17:738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Tham D, Wei W, Shin YS, Ma C, Ahmad H, Shi Q, Yu J, Levine RD, Heath JR. Nano Lett. 2012;12(12):6101–6106. doi: 10.1021/nl302748q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei W, Shin YS, Mao C, Wang J, Elitas M, Fan R, Heath JR. Genome Medicine. 2013;5:75. doi: 10.1186/gm479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma C, Fan R, Elitas M. Front Oncol. 2013;3:133–7. doi: 10.3389/fonc.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chattopadhyay PK, Gierahn TM, Roederer M, Lobe JC. Nat Imm. 2014;15:128–135. doi: 10.1038/ni.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin LD, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S. Development. 2009;136:3853–3862. doi: 10.1242/dev.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Zhou J, Sutherland A, Wei W, Shin YS, Xue M, Heath JR. Ann Rev Anal Chem. 2014:15–53. doi: 10.1146/annurev-anchem-071213-020323. [DOI] [PubMed] [Google Scholar]
- 32.Chen MB, Whisler JA, Jeon JS, Kamm RD. Integr Biol. 2013;5:1262–1271. doi: 10.1039/c3ib40149a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.