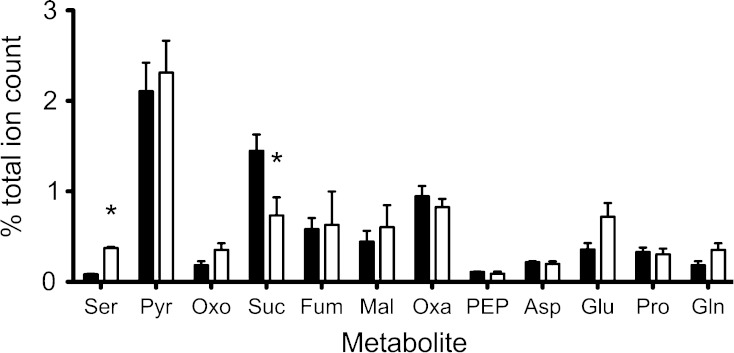
Fig. 2.
Key metabolite differences between C. jejuni NCTC 11168 and sdaA strains. Following ESI-TOF MS of five biological replicates in negative ionisation mode, m/z peaks were assigned putative metabolite identities to an accuracy of 0.2 Da for C. jejuni NCTC 11168 (black bars) and sdaA strains (open bars). Shown are those metabolites detected that are most closely related to amino-acid catabolism [Ser serine (104.00); Pyr pyruvate (87.00); Oxo 2-oxoglutarate (145.00); Suc Succinate (117.00); Fum Fumarate (115.00); Mal Malate (133.00); Oxa oxaloacetate (134.00); PEP phosphoenolpyruvate (167.00); Asp aspartate (132.00); Glu glutamate (146.00); Pro proline (115.00); Gln glutamine (145.00)] as a % of the total ion count. * p value <0.05 between C. jejuni NCTC 11168 and sdaA strain