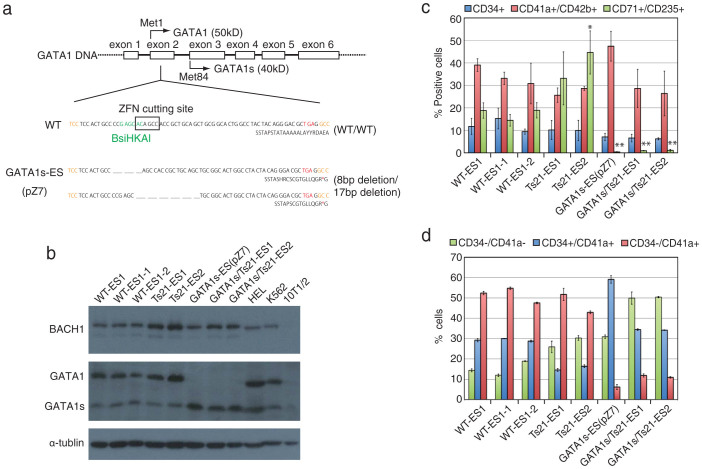
Figure 2. Characterisation of GATA1s-ES and GATA1s/Ts21-ES cells.
(a) Sequence analyses of GATA1s-ES cells. One allele had an 8-bp deletion and the other had a 17-bp deletion; both resulted in a TGA stop codon in exon 2 of GATA1. The GATA1 nucleotide (upper line) and amino acid (lower line) sequences are shown for WT-ES and GATA1s-ES cells. An asterisk shows the stop codon. (b) Western blot analyses of erythroid lineage-differentiated cells derived from WT-ES, WT-ES sublines, Ts21-ES lines, GATA1s-ES, and GATA1s/Ts21-ES lines. Anti-BACH1 was used to detect the gene-dosage effect on hChr.21. Anti-GATA1 recognised the C-terminus of both GATA1 and GATA1s protein. Anti-α-tubulin was used as an internal control. HEL cell lysate and K562 nuclear extract were used as positive controls. 10T1/2 whole cell lysate was used as a negative control. Cropped blots were used in this figure. Original full-length blots are shown in supplementary figure 12. (c, d) Haematopoietic differentiation analyses using WT-ES, WT-ES sublines, Ts21-ES lines, GATA1s-ES, and GATA1s/Ts21-ES lines. Data are the means of 3 independent experiments (±S.D.). The percentage of CD34+, CD41a+/CD42b+, and CD71+/CD235+ cells are shown in each differentiation stage (ES-sac (day 14), megakaryocyte (day 20), and erythroid (day 20)) (c). Statistical analyses were performed by comparison with WT-ES cells (WT-ES1, WT-ES1-1 and WT-ES1-2). *p < 0.05, **p < 0.01 by two-tailed Student's t test. The percentage of CD34-/CD41a-, CD34+/CD41a+ and CD34-/CD41a+ cells are shown in the megakaryocyte stage (d).