Abstract
Aims
Small cell lung cancer (SCLC) carries a poor prognosis, and the systemic therapies currently used as treatments are only modestly effective, as demonstrated by a low 5-year survival at only ∼5%. In this retrospective collected from March 2013 to study, we performed comprehensive genomic profiling of 98 small cell undifferentiated lung cancer (SCLC) samples to identify potential targets of therapy not currently searched for in routine clinical practice.
Methods
DNA from 98 SCLC was sequenced to high, uniform coverage (Illumina HiSeq 2500) and analysed for all classes of genomic alterations.
Results
A total of 386 alterations were identified for an average of 3.9 alterations per tumour (range 1–10). Fifty-two (53%) of cases harboured at least 1 actionable alteration with the potential to personalise therapy including base substitutions, amplifications or homozygous deletions in RICTOR (10%), KIT (7%), PIK3CA (6%), EGFR (5%), PTEN (5%), KRAS (5%), MCL1 (4%), FGFR1 (4%), BRCA2, (4%), TSC1 (3%), NF1 (3%), EPHA3 (3%) and CCND1. The most common non-actionable genomic alterations were alterations in TP53 (86% of SCLC cases), RB1 (54%) and MLL2 (17%).
Conclusions
Greater than 50% of the SCLC cases harboured at least one actionable alteration. Given the limited treatment options and poor prognosis of patients with SCLC, comprehensive genomic profiling has the potential to identify new treatment paradigms and meet an unmet clinical need for this disease.
Keywords: Lung Cancer, Molecular Pathology, Molecular Oncology, Molecular Genetics
Small cell lung cancer (SCLC) is a well-recognised histologic variant of lung cancer with a distinct histologic appearance and unique biology.1–4 SCLC is a neuroendocrine carcinoma with neurosecretory granules identified in the scant tumour cytoplasm on electron microscopy and positive immunostaining for neuropeptide antigens such as synaptophysin and chromogrannin.1–4 SCLC accounts for approximately 16–18% of all newly diagnosed lung cancers in the USA which translates into approximately 30 000 new cases each year.5 In comparison with non-small cell lung cancer (NSCLC), SCLC features a shorter doubling time, higher growth fraction, earlier development of widespread metastases, and strong 60–80% initial response rate to etoposide-based chemotherapy and radiation treatment.1–5 However, the majority of SCLC patients suffer relapse of the disease within 3–6 months after cessation of therapy and feature a mean overall of just 6 months from the time of relapse.5 For patients who do not respond to the front line chemotherapy, the overall survival is worse averaging only approximately 6 months from the time of diagnosis.1–5 Moreover, the 5-year survival rate for all SCLC cases is only 5%.1–5 As opposed to other types of primary lung cancer, most notably lung adenocarcinoma, well-defined genomic alterations and opportunities for targeted therapy for SCLC have not, to date been identified. We hypothesised that comprehensive genomic profiling of clinical SCLC samples by next generation sequencing (NGS) could identify genomic-derived drug targets of therapy for patients diagnosed with this aggressive malignancy in a single diagnostic test.
Methods
Hybridisation-based capture of 3320 exons from 182 cancer-related genes and 37 introns of 14 genes commonly rearranged in cancer (previous version of the test) and 3769 exons from 236 cancer-related genes and 47 introns of 19 genes commonly rearranged in cancer (current version of the test) was applied to ≥50 ng of DNA extracted from 98 SCLC tumour specimens and sequenced to high, uniform coverage with a mean sequencing depth of 714× as previously described.6 Consistent median sequencing depth was achieved by processing specimens according to optimised, locked down, standard operating procedures (SOP) on automated liquid handlers in a Clinical Laboratory Improvement Act (CLIA)-certified laboratory as previously described.6 The study population consisted of 98 consecutive cases of SCLC collected from March, 2013 through February, 2014 for which tumour samples were submitted to Foundation Medicine for NGS assessment. Genomic alterations (base substitutions, small indels, rearrangements, copy number alterations) were determined and then reported for these patient samples. Actionable genomic alteration (GA) were defined as those identifying anticancer drugs on the market or in registered clinical trials. Local site permissions to use clinical samples were used for this study.
There were 60 female and 38 male SCLC patients (see online supplementary table S1) with a median age 60.7 years (range 35–82 years). By definition, all (100%) tumours were high grade of which 1 (1%) stage I, 2 (2%) stage II, 22 (23%) stage III and 72 (74%) stage IV tumours at the time of sequencing. The tissue samples sequenced originated from a biopsy of the primary SCLC in 48 (49%) of cases and from the following metastatic tumour sites in 50 (51%): regional lymph nodes (19 cases), liver (12 cases), brain (4 cases), pleura/pleural fluid (4 cases), chest wall (2 cases), mediastinum (2 cases), head and neck (2 cases), and 1 case each from the spine, trachea, small intestine, bone and soft tissue. The relative percentage of tumour cell nuclear area to benign stromal/non-tumoral nuclear area varied from a low of 20% to a high of 90%, with a mean of 52%.
Results
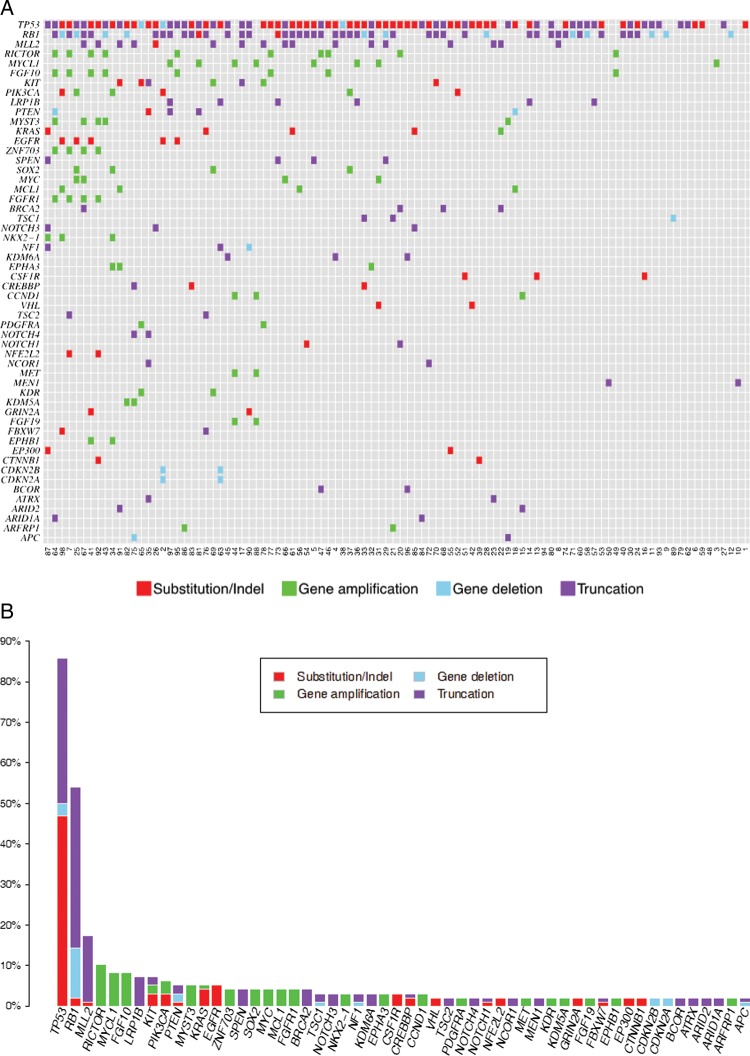
All 98 SCLC (100%) specimens harboured at least one genomic alteration with 386 total alterations identified for an average of 3.9 alterations per tumour (range 1–10). There were no observed differences in the quality of the sequencing results among the multiple different types of tissue samples used for DNA extraction. Of the 386 total alterations, there were 200 base substitutions, 55 short insertions and deletions, 99 gene amplifications, 26 homozygous deletions and 6 rearrangements/fusions. Ninety-six (25%) alterations were considered to be actionable with the potential to personalise targeted treatment. Fifty-two (53%) of cases harboured at least one actionable alteration (0.98 actionable alterations per the entire cohort of SCLC) including base substitutions, amplifications or homozygous deletions in RICTOR (10%), KIT (7%), PIK3CA (6%), EGFR (5%), PTEN (5%), KRAS (5%), MCL1 (4%), FGFR1 (4%), BRCA2, (4%), TSC1 (3%), NF1 (3%), EPHA3 (3%) and CCND1 (3%) (figure 1A,B). Of the seven most commonly altered genes, only one gene (RICTOR) was considered to be actionable (table 1). The most common non-actionable genomic alterations were alterations in TP53 (86% of SCLC cases), RB1 (54%) and MLL2 (17%).
Figure 1.
Genomic alterations in small cell undifferentiated lung cancer. (A) is the tile plot showing the alterations in each individual tumour. (B) is the bar plot showing the absolute and relative frequency of each alteration found in the study.
Table 1.
Twenty-nine most frequently altered genes in 98 cases of SCLC
| Gene | Predicted to be actionable | Substitution/indel | Amp | Deletion | Truncation | Fusion/rearrangement | Number of samples | Percentage of samples (%) |
|---|---|---|---|---|---|---|---|---|
| TP53 | No | 46 | 0 | 3 | 35 | 0 | 84 | 86 |
| RB1 | No | 2 | 0 | 12 | 39 | 0 | 53 | 54 |
| MLL2 | No | 1 | 0 | 0 | 16 | 0 | 17 | 17 |
| RICTOR | Yes | 0 | 10 | 0 | 0 | 0 | 10 | 10 |
| MYCL1 | No | 0 | 8 | 0 | 0 | 0 | 8 | 8 |
| FGF10 | No | 0 | 8 | 0 | 0 | 0 | 8 | 8 |
| LRP1B | No | 0 | 0 | 0 | 7 | 0 | 7 | 7 |
| KIT | Yes | 3 | 2 | 0 | 2 | 0 | 7 | 7 |
| PIK3CA | Yes | 3 | 3 | 0 | 0 | 0 | 6 | 6 |
| PTEN | Yes | 1 | 0 | 2 | 2 | 0 | 5 | 5 |
| MYST3 | No | 0 | 5 | 0 | 0 | 0 | 5 | 5 |
| KRAS | Yes | 4 | 1 | 0 | 0 | 0 | 5 | 5 |
| EGFR | Yes | 5 | 0 | 0 | 0 | 0 | 5 | 5 |
| ZNF703 | No | 0 | 4 | 0 | 0 | 0 | 4 | 4 |
| SPEN | No | 0 | 0 | 0 | 4 | 0 | 4 | 4 |
| SOX2 | No | 0 | 4 | 0 | 0 | 0 | 4 | 4 |
| MYC | No | 0 | 4 | 0 | 0 | 0 | 4 | 4 |
| MCL1 | Yes | 0 | 4 | 0 | 0 | 0 | 4 | 4 |
| FGFR1 | Yes | 0 | 4 | 0 | 0 | 0 | 4 | 4 |
| BRCA2 | Yes | 0 | 0 | 0 | 4 | 0 | 4 | 4 |
| TSC1 | Yes | 0 | 0 | 1 | 2 | 0 | 3 | 3 |
| NOTCH3 | No | 0 | 0 | 0 | 3 | 0 | 3 | 3 |
| NKX2-1 | No | 0 | 3 | 0 | 0 | 0 | 3 | 3 |
| NF1 | Yes | 0 | 0 | 1 | 2 | 0 | 3 | 3 |
| KDM6A | No | 0 | 0 | 0 | 3 | 0 | 3 | 3 |
| EPHA3 | Yes | 0 | 3 | 0 | 0 | 0 | 3 | 3 |
| CSF1R | No | 3 | 0 | 0 | 0 | 0 | 3 | 3 |
| CREBBP | No | 2 | 0 | 0 | 1 | 0 | 3 | 3 |
| CCND1 | Yes | 0 | 3 | 0 | 0 | 0 | 3 | 3 |
SCLC, Small cell lung cancer.
Discussion
The known genomic landscape of SCLC classically features high frequencies of RB1 and TP53 mutation which were recapitulated in this study.7–10 Additional alterations identified in this study involve a wide variety of recognised cancer-related genes, and impact a series of genomic pathways that have been previously linked to development and progression of SCLC.7–10 By comparison with other solid tumour types, including NSCLC,11 the frequency of potentially actionable genomic alterations in SCLC is lower with an average of 0.98 actionable GA per patient. Around 50% of the SCLC patients in this series harboured at least one actionable genomic alteration. Not only does SCLC feature a lower frequency of actionability than other types of lung cancer such as adenocarcinoma,11 the long tail of altered genes in SCLC and the resulting wide panorama of impacted mechanisms of tumour biology are highly complex. Additionally, given that only 25% of the altered genes in this series of SCLC cases are currently considered to be actionable, it is critical that the sequencing test used to assess the tumours for potential therapy targets be sensitive enough not to miss any of these important alterations. This finding necessitates that a broad diagnostic assay that can detect these genomic changes at a high degree of sensitivity from limited biopsy material be used to maximise targeted therapeutic options in an individual patient.
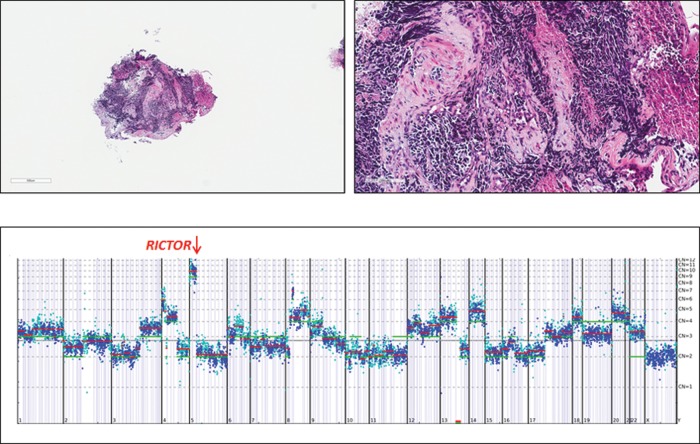
Of the seven most frequently altered genes in SCLC, the only potentially actionable gene in this group of alterations is RICTOR amplification which was found in 10 (10%) of SCLC cases (figure 1A,B). All 10 (100%) of the alterations of RICTOR in this study of SCLC were amplifications. An example of RICTOR amplification in SCLC is seen in case 45 (figure 2). RICTOR encodes the protein, RICTOR (rapamycin-insensitive companion of mTOR), an mTOR binding protein that interacts with mTOR in the complex mTORC2.12 When all types of NSCLC are included, amplification of RICTOR has been found in 8–10% of cases, but there is no data for the frequency of RICTOR amplification in SCLC currently available (cBioPortal for Cancer Genomics, Oct 2013). Tumours with RICTOR amplification may be sensitive to inhibitors of mTORC2, the RICTOR-containing complex.13 Numerous inhibitors that target both mTORC1 and mTORC2 complexes, as well as dual PI3K/mTOR inhibitors, are under preclinical and clinical investigation in multiple tumour types.14 15
Figure 2.
Bronchial biopsy from a 78-year-old man demonstrating classic histology for small cell undifferentiated carcinoma (case number 45). The low magnification image (1×) of the tumour is at the upper left and the high magnification image (20×) at upper right. The tumour was stage IV at the time of diagnosis with extensive bone metastases. The tumour was sequenced to a mean coverage depth of 610× and NGS revealed an amplification of the RICTOR gene at 13 copies shown in the lower portion of the figure. There were also lower level amplifications of FGFR3, FGF10 and MYST3. The tumour also had base substitutions in TP53 (G266*) and MSH6 (V509A) along with a loss (homozygous deletion) in RB1. The chromosomal location is provided in the X axis below and the gene copy number on the Y axis to the right of the gene copy number plot.
Additional potentially actionable alterations in this series of SCLC involved the KIT (multiple types of alterations in 7% of SCLC) and EGFR (base substitutions in 5% of SLCL) genes. KIT mutations in SCLC are rare and have been reported only in 2.1% in the COSMIC database (COSMIC, January 2014). KIT protein expression has been reported in 36.4–83.3% of SCLC samples.16 17 A number of tyrosine kinase inhibitors that target KIT have been successful for patients with various KIT-mutated solid tumours. Additionally, PI3K inhibitors and mTOR inhibitors, may have potential for treatment of a tumour with either a KIT amplification or activating mutation. The combination of first-line kinase inhibitors with MEK, PI3K, or mTOR inhibitors, or new therapies such as switch kinase inhibitors, may be a useful strategy to target kinase inhibitor-resistant tumours. EGFR mutations have been reported in 2–5% of SCLC (COSMIC, February 2014).18 19 Although activating mutations in EGFR have been shown to confer sensitivity to EGFR tyrosine kinase inhibitors such as erlotinib, gefitinib and afatinib in NSCLC,20–23 studies showing responsiveness to gefitinib or erlotinib in EGFR-mutated SCLC have been limited.24 25
SOX2 amplification was detected in 4% of SCLC in the current study. SOX2 encodes SOX2, a transcription factor described as a ‘lineage survival’ oncogene and SOX2 expression may be associated with resistance to cytotoxic chemotherapy.26 This result is in contrast with the previously reported frequency of high-level SOX2 amplification which has previously been found in 27% (15/56) SCLC.27 The presence of SOX2 expression does not appear to relate to prognosis or survival in patients with SCLC.28 There are currently no therapies available to directly target SOX2 amplification in cancer. However, preclinical research suggests that SOX2 expression may predict sensitivity to inhibitors of Cdk4 and Cdk6.29
In summary, high-sensitivity genomic profiling can discover potential new routes to targeted therapies in patients with SCLC who have relapsed after primary chemotherapy. Given the well-known poor prognosis for relapsed SCLC, further study of the detection of genomic alterations and the potential for targeted therapies to help these patients appears warranted.
Take-home messages.
A sensitive/validated next-generation sequencing assay can readily be performed on formalin-fixed paraffin embedded biopsies of patients diagnosed with small cell undifferentiated carcinoma of the lung.
Fifty-two (53%) of small cell lung cancer (SCLC) cases harboured at least one actionable alteration with the potential to personalise therapy including base substitutions, amplifications or homozygous deletions in RICTOR (10%), KIT (7%), PIK3CA (6%), EGFR (5%), PTEN (5%), KRAS (5%), MCL1 (4%), FGFR1 (4%), BRCA2, (4%), TSC1 (3%), NF1 (3%), EPHA3 (3%) and CCND1.
High-sensitivity genomic profiling can discover potential new routes to targeted therapies in patients with SCLC who have relapsed after primary chemotherapy.
Supplementary Material
Footnotes
Contributors: The following authors contributed to the development and submission of this manuscript as follows: substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; drafting the work or revising it critically for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: JSR, KW, ORE, AT, LF, CES, GAO, GP, RY, DL, JC, SMA, JAE, DM, VAM and PJS.
Competing interests: None.
Ethics approval: Patient identity protection was maintained throughout the study which was considered to be an exempt study by the Institutional Review Board of the Albany Medical College, Albany, NY USA and Ethics Committee at Foundation Medicine, Inc., Cambridge, MA USA.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Additional data for this study is provided in online supplementary table S1.
References
- 1.Cooper S, Spiro SG. Small cell lung cancer: treatment review. Respirology 2006;11:241–8 [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sørensen M, Pijls-Johannesma M, Felip E; ESMO Guidelines Working Group. Small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010;21(Suppl 5):v120–5 [DOI] [PubMed] [Google Scholar]
- 4.van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet 2011;378:1741–55 [DOI] [PubMed] [Google Scholar]
- 5.Hanna N, Bunn PA, Jr, Langer C, et al. Randomized phase III trial comparing irinotecan/cisplatin with etoposide/cisplatin in patients with previously untreated extensive-stage disease small-cell lung cancer. J Clin Oncol 2006;24:2038–43 [DOI] [PubMed] [Google Scholar]
- 6.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahel RA, Weber E. Small cell lung cancer: the new biology. Semin Radiat Oncol 1995;5:11–18 [DOI] [PubMed] [Google Scholar]
- 8.D'Angelo SP, Pietanza MC. The molecular pathogenesis of small cell lung cancer. Cancer Biol Ther 2010;10:1–10 [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Wannesson L. A genetic snapshot of small cell lung cancer. Cancer Discov 2012;2:769–71 [DOI] [PubMed] [Google Scholar]
- 10.Stovold R, Blackhall F, Meredith S, et al. Biomarkers for small cell lung cancer: neuroendocrine, epithelial and circulating tumour cells. Lung Cancer 2012;76:263–8 [DOI] [PubMed] [Google Scholar]
- 11.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber JD, Gutmann DH. Deconvoluting mTOR biology. Cell Cycle 2012;11:236–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sparks CA, Guertin DA. Targeting mTOR: prospects for mTOR complex 2 inhibitors in cancer therapy. Oncogene 2010;29:3733–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest 2011;121:1231–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schenone S, Brullo C, Musumeci F, et al. ATP-competitive inhibitors of mTOR: an update. Curr Med Chem 2011;18:2995–3014 [DOI] [PubMed] [Google Scholar]
- 16.López-Martin A, Ballestín C, Garcia-Carbonero R, et al. Prognostic value of KIT expression in small cell lung cancer. Lung Cancer 2007;56:405–13 [DOI] [PubMed] [Google Scholar]
- 17.Lu HY, Zhang G, Cheng QY, et al. Expression and mutation of the c-kit gene and correlation with prognosis of small cell lung cancer. Oncol Lett 2012;4:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatematsu A, Shimizu J, Murakami Y, et al. Epidermal growth factor receptor mutations in small cell lung cancer. Clin Cancer Res 2008;14:6092–6 [DOI] [PubMed] [Google Scholar]
- 19.Shiao TH, Chang YL, Yu CJ, et al. Epidermal growth factor receptor mutations in small cell lung cancer: a brief report. J Thorac Oncol 2011;6:195–8 [DOI] [PubMed] [Google Scholar]
- 20.Roengvoraphoj M, Tsongalis GJ, Dragnev KH, et al. Epidermal growth factor receptor tyrosine kinase inhibitors as initial therapy for non-small cell lung cancer: focus on epidermal growth factor receptor mutation testing and mutation-positive patients. Cancer Treat Rev 2013;39:839–50 [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi K, Hagiwara K. Epidermal growth factor receptor (EGFR) mutation and personalized therapy in advanced nonsmall cell lung cancer (NSCLC). Target Oncol 2013;8:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi A, Remick S, Tse W. EGFR inhibition in non-small cell lung cancer: current evidence and future directions. Biomark Res 2013;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keating GM. Afatinib: a review of its use in the treatment of advanced non-small cell lung cancer. Drugs 2014;74:207–21 [DOI] [PubMed] [Google Scholar]
- 24.Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006;355:213–15 [DOI] [PubMed] [Google Scholar]
- 25.Lu HY, Wang XJ, Mao WM. Targeted therapies in small cell lung cancer. Oncol Lett 2013;5:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsunoda Y, Sakamoto M, Sawada T, et al. Characteristic genes in luminal subtype breast tumors with CD44+CD24-/low gene expression signature. Oncology 2011;81:336–44 [DOI] [PubMed] [Google Scholar]
- 27.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S, Xu Y, Chen Y, et al. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PLoS ONE 2012;7:e36326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flaherty KT, Lorusso PM, Demichele A, et al. Phase I, dose-escalation trial of the oral cyclin-dependent kinase 4/6 inhibitor PD 0332991, administered using a 21-day schedule in patients with advanced cancer. Clin Cancer Res 2012;18:568–76 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.