Abstract
Endocrine disrupting chemicals (EDCs) including plasticizers, pesticides, detergents and pharmaceuticals, affect a variety of hormone-regulated physiological pathways in humans and wildlife. Many EDCs are lipophilic molecules and bind to hydrophobic pockets in steroid receptors, such as the estrogen receptor and androgen receptor, which are important in vertebrate reproduction and development. Indeed, health effects attributed to EDCs include reproductive dysfunction (e.g., reduced fertility, reproductive tract abnormalities and skewed male/female sex ratios in fish), early puberty, various cancers and obesity. A major concern is the effects of exposure to low concentrations of endocrine disruptors in utero and post partum, which may increase the incidence of cancer and diabetes in adults. EDCs affect transcription of hundreds and even thousands of genes, which has created the need for new tools to monitor the global effects of EDCs. The emergence of massive parallel sequencing for investigating gene transcription provides a sensitive tool for monitoring the effects of EDCs on humans and other vertebrates as well as elucidating the mechanism of action of EDCs. Zebrafish conserve many developmental pathways found in humans, which makes zebrafish a valuable model system for studying EDCs especially on early organ development because their embryos are translucent. In this article we review recent advances in massive parallel sequencing approaches with a focus on zebrafish. We make the case that zebrafish exposed to EDCs at different stages of development, can provide important insights on EDC effects on human health.
Keywords: endocrine disruptors, steroid receptors, zebrafish, massive parallel sequencing
ENDOCRINE DISRUPTORS
There is increasing concern about the effects on the health of humans and wild life of organic and inorganic contaminants that have been introduced into the environment by manufacturing industries (Baker, et al. 2012; Baker and Chandsawangbhuwana 2012; Calafat, et al. 2008; Colborn, et al. 1993; Diamanti-Kandarakis, et al. 2009; Heindel and vom Saal 2009; Rubin 2011). Many of these chemicals are used as plasticizers, pesticides, detergents and pharmaceuticals providing important benefits for modern industrial societies. However, some of these chemicals disrupt a variety of hormone-regulated physiological pathways, including reproductive responses mediated by the estrogen receptor [ER] and androgen receptor [AR] in humans (Calafat et al. 2008; Diamanti-Kandarakis et al. 2009; Kelce, et al. 1998; Rubin 2011; Sonnenschein and Soto 1998; Soto, et al. 2009; Soto, et al. 2008; Swan 2008) and wildlife (Lange, et al. 2009; Oehlmann, et al. 2008; Oehlmann, et al. 2009). In humans exposure to endocrine disrupting chemicals [EDCs] may lead to premature puberty in females, and decreased reproductive ability in men. Of major concern is transient exposure to EDCs in utero and in newborns, which can have toxic effects on reproduction and development, as well as causing some endocrine-related cancers and heart disease later in life (Grun and Blumberg 2006; Henley and Korach 2006; Soto et al. 2008; Swan 2008).
Estrogens, androgens and other steroids [Figure 1] and many EDCs [Figure 2] are small molecules, lipophilic in nature, which is thought to explain binding of EDCs to the ER, AR and other steroid receptors. For example, chemicals, such as 4-nonylphenol [4-NP], which contains a phenolic group that mimics the A ring on estradiol [E2], and bisphenol A [BPA], which mimics the A and D rings on E2, can disrupt physiological responses mediated by the ER (Baker and Chandsawangbhuwana 2012; Diamanti-Kandarakis et al. 2009; Rubin 2011; Sonnenschein and Soto 1998).
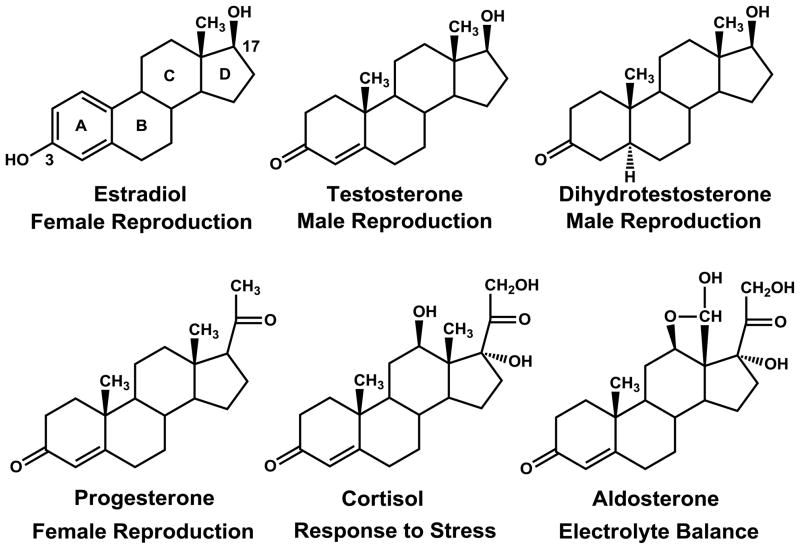
Figure 1. Adrenal and Sex Steroids.
Estradiol [E2] is the canonical female sex steroid. However, E2 is more than a reproductive steroid: E2 has important actions in heart, brain, liver, bone in females and males (Baker 2013; Gao and Dahlman-Wright 2011; Heldring, et al. 2007; Sugiyama, et al. 2010). Moreover, E2 has an important role in prostate physiology (Prins, et al. 2011; Weihua, et al. 2002). Thus, EDC binding to the ER can affect reproductive physiology in males as well as non-reproductive physiology in males and females. Testosterone [T] and 5α-dihydrotestosterone [DHT] are two male androgens (Sharifi and Auchus 2012), which also are important hormones for females.
In humans, progesterone is important for successful implantation of the fertilized egg (Graham and Clarke 1997; Smith 2007). Progesterone antagonists, such as RU486, can interfere with implantation and prevent pregnancy. However, as with E2, progesterone has actions in males and in tissues that are not directly involved in reproduction.
Cortisol is involved in response to stress, metabolism of carbohydrates and lipids, bone turnover, lung maturation, and homeostasis of the immune, cardiovascular and central nervous systems (McEwen 2012; Odermatt and Gumy 2008; Sapolsky, et al. 2000; Tomlinson, et al. 2004; Zhou and Cidlowski 2005). Although these actions are mediated primarily by the glucocorticoid receptor [GR], in some cells, cortisol is a transcriptional activator of the mineralocorticoid receptor [MR].
Aldosterone regulates electrolyte homeostasis by controlling transport of sodium and potassium in kidney and gut through transcriptional activation of the MR (Hawkins, et al. 2012; Martinerie, et al. 2013). However, MR also is important in other organs such as heart and brain, in which the MR may be activated by aldosterone or cortisol (Funder 2009; Tomlinson et al. 2004).
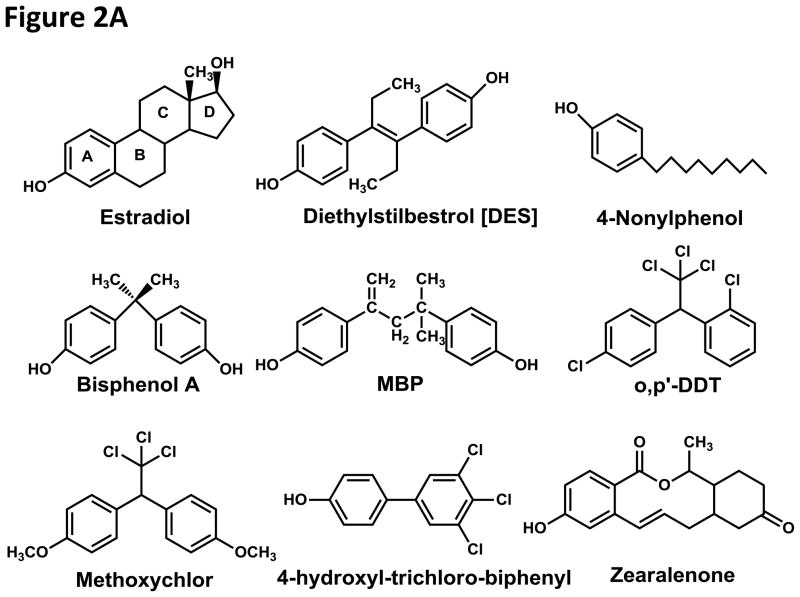
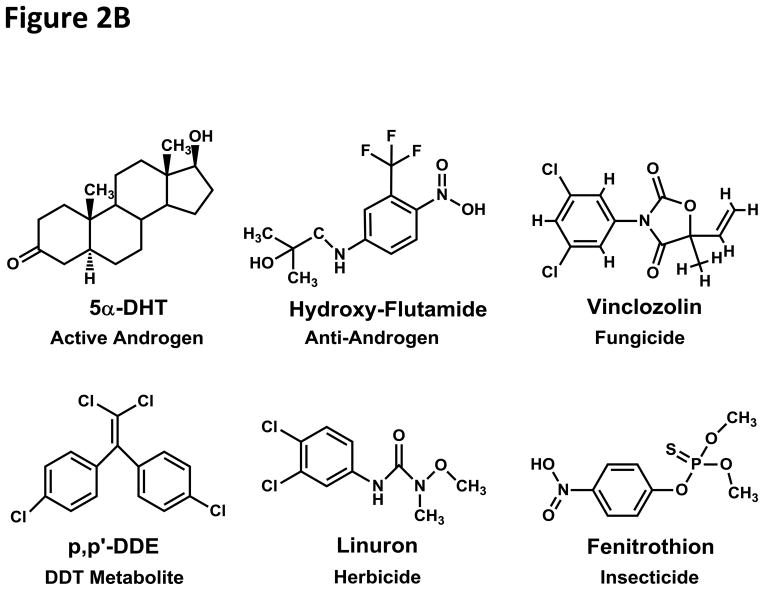
Figure 2. Structures of Xenoestrogens and Xenoandrogens.
A. Xenoestrogens (Diamanti-Kandarakis et al. 2009; Kuiper, et al. 1998; Sonnenschein and Soto 1998) A key property of xenoestrogens is the presence of one or more phenolic groups in a structure that mimics either A or the A and D rings of E2. Examples are diethylstilbestrol, [DES], a synthetic estrogen, 4-nonylphenol [NP] and bisphenol A [BPA], which are used in plastics, 4-methyl-2,4-bis(p-hydroxyphenyl)pent-1-ene (MBP), a metabolite of BPA (Baker and Chandsawangbhuwana 2012; Yamaguchi, et al. 2005), which has over 100-fold higher affinity for the ER than BPA and zearalenone, which is a toxic estrogenic metabolite synthesized by some fungi (Katzenellenbogen, et al. 1979). However, chemicals without hydroxyl substituents also can disrupt estrogen physiology. Examples are DDT (Sonnenschein and Soto 1998), a pesticide, and polychlorinated hydroxybiphenyls [PCBs], which are used in transformers and capacitors (Korach, et al. 1988; Kuiper et al. 1998).
B. Xenoandrogens (Diamanti-Kandarakis et al. 2009; Kelce et al. 1998; Luccio-Camelo and Prins 2011; Sonnenschein and Soto 1998). A variety of structures, which do not have much in common with either DHT or T can bind the AR. Vinclozolin, a fungicide, is an antiadrogen, as are p,p′-DDE, a metabolite of p,p′-DDT, the insecticide fenitrothion (Tamura, et al. 2001) and the herbicide linuron (Lambright, et al. 2000). Hydroxy-flutamide is an anti-androgen used in treating prostate cancer.
The ER, AR and other steroid receptors are nuclear receptors, a large and diverse family of transcription factors (Baker 2005; Bertrand, et al. 2011; Bertrand, et al. 2004; Escriva, et al. 2000; Krasowski, et al. 2011; Markov, et al. 2009; Sladek 2011) that are activated by a variety of lipophilic molecules (Baker 2011; Chawla, et al. 2001; Huang, et al. 2010; Ingraham and Redinbo 2005; Markov et al. 2009; Sladek 2011). Nuclear receptors regulate differentiation, development, homeostasis and responses to stress in humans and other vertebrates. Thus, disruption of nuclear receptor signaling by EDCs poses a threat to many physiological responses in humans, fish and other animals (Diamanti-Kandarakis et al. 2009; Grun and Blumberg 2006; Heindel and vom Saal 2009; Nelson, et al. 2013; Soto et al. 2009; Swan 2008; Vandenberg, et al. 2009). However, nuclear receptors are not the only target for EDCs. As more and more diverse chemicals are synthesized and dispersed into the environment, it is likely that additional physiological responses will be disrupted. As a result, government agencies and environmental scientists have a critical need for methods to monitor both short and long term effects of exposure to low concentrations of EDCs on human health.
ZEBRAFISH: A MODEL FOR STUDYING ENDOCRINE DISRUPTION
Zebrafish [Danio rerio] have emerged as an important tool for studying the biological effects of hormones and EDCs. Zebrafish is a small tropical fresh-water fish indigenous to northern India, northern Pakistan, Nepal, and Bhutan in South Asia. Zebrafish are relatively inexpensive to maintain compared to rats and mice and are easily bred in large numbers. Zebrafish have short life spans [60 days to maturity]. Thus, three generations of zebrafish can be easily generated within a twelve month period, making it possible to carry out immediate studies in response to exposure to endocrine disruptors, in addition to longitudinal transgenerational studies analysis with F1 and F2 progeny derived from the exposed fish. Zebrafish embryos are optically transparent, allowing microscopic observations of organs as they develop (Lieschke and Currie 2007; Segner 2009). Thus, one can study the onset and course of exposure to EDC s on organ development in vivo and in real time. At this point there is no marine vertebrate model that is as well characterized as the zebrafish. The highly inbred, lab-dependent nature of these animals and the availability of the complete genome sequence make them an ideal model for toxicology studies and rapid phenotypic assessment (Lieschke and Currie 2007).
Zebrafish genome
The draft genome of the zebrafish genome has been publicly available for over a decade, facilitating many novel discoveries, including positional cloning of hundreds of genes from mutations. Recent sequencing efforts using massively parallel sequencing have focused on generating a high-quality reference genome similar to those available for the human and mouse genomes. The Zv9 assembly reported recently by Howe and colleagues is a hybrid of high-quality finished clone sequence (83%) and whole-genome shotgun (WGS) sequence (17%), with a total size of 1.4123 gigabases (Gb) (Howe, et al. 2013). This reference sequence is linked to a high-resolution, high-density meiotic map. It greatly facilitates the identification of mutations, because it enables direct comparison of both mutated and normal sequences (Schier 2013). Analysis of the reference has revealed that more than 75% of human genes implicated in disease have counterparts in zebrafish. An advantage of the annotated reference genome is possibilities for exome-enrichment deep sequencing experiments analogous to those now routinely performed with human genome samples. This will accelerate positional cloning projects and novel genome-wide mutation discovery efforts (Howe et al. 2013). This further highlights the utility of the zebrafish model for elucidation of gene function and studies of the effects of endocrine disruption.
Genetic modification
Zebrafish embryos and adults can be genetically modified by microinjection, chemical mutagenesis and transgenesis. Moreover, the zebrafish database ZFIN [http://zfin.org] (Bradford, et al. 2011) provides up-to-date information on all aspects of zebrafish. Most important, many of the developmental processes in zebrafish are conserved in humans (Barbazuk, et al. 2000; Dooley and Zon 2000; Woolfe, et al. 2004). The relevance to humans of many genes and pathways for development in zebrafish makes these fish an excellent system to study possible effects of EDCs on human health. In addition, data from zebrafish exposed to samples taken from sewage outfalls (Lange et al. 2009; Vajda, et al. 2008), rivers, drinking water, soil, food, etc. can be used to determine their toxicity to humans of EDCs from different sources.
An ere-zvtg1: gfp transgenic zebrafish line for environmental monitoring of endocrine disrupters was developed recently for environmental monitoring of endocrine disrupters. In this transgenic model under control conditions, green fluorescent protein (GFP) was exclusively expressed in the liver of mature adult female fish. Male and larval transgenic fish did not express GFP but could be induced to express GFP in the liver after exposure to 17-alpha-ethynylestradiol (EE2) (Chen, et al. 2010; Gorelick and Halpern 2011; Lee, et al. 2012)
MONITORING EXPOSURE TO ENDOCRINE DISRUPTION
Monitoring pollution at environmental sites has typically involved, sampling sediment or water from that location, determination of the constituents present and comparison of the levels of contaminants to reference levels associated with adverse exposure outcomes. In many cases only a percentage of contaminants are bio-available making this approach problematic. Further confounding this issue is the synergistic effects provided by compound mixtures, which can yield outcomes that differ significantly from single compound exposures. There is a growing need to develop alternate approaches to biomonitoring which encompass chemical analysis and bioavailability studies of pollutants end point phenotypic and biological assays (Celiz, et al. 2009; Letcher, et al. 2010).
An advantage of using zebrafish to monitor exposure to EDCs is that an oral method can be used to administer EDCs to zebrafish. Zebrafish are raised in glass aquaria, at a constant temperature of 29 °C with a light-dark cycle ratio of 14:10 hours. The fish typically are acclimated for one week prior to commencing the exposure experiments. EDC exposures utilize a continuous flow-through system, to maintain constant concentrations of the chemicals during the duration of the exposure, after which the fish can be selected for phenotypic and genomic analyses. Recent advances in molecular methods, such as microarrays, have substantially improved the sensitivity for assessing the effects of EDCs. Indeed, the ability to screen a wide range of endocrine responses in organisms from polluted waters and monitor harmful effluents entering the ecosystem is facilitated by microarray technology.
DNA Microarray Technology
The multiplicity of genes that will have altered transcription in humans and wildlife due to exposure to EDCs requires comprehensive assays for analyzing gene transcription. DNA microarrays have been useful for this purpose (Brown and Botstein 1999; Hardiman 2004). Microarrays facilitate measurement of expression levels of hundreds to thousands of genes simultaneously from a single tissue sample. This technology has been widely adopted by the biomedical research community and for the past decade it has been a key molecular tool in basic, translational, and clinical research. Microarray technology has found widespread use by the zebrafish community, in applications as diverse as rod photoreceptor degeneration and regeneration (Morris, et al. 2011), the toxic effects of nanoparticulate silver in zebrafish (Griffitt, et al. 2013), and differences in response to teratogenic and non-teratogenic exposure concentrations of bisphenol A and 17beta-estradiol (Saili, et al. 2013). Although their primary use has been for gene expression analyses, microarrays have also found utility in genotyping and re-sequencing applications (Hardiman 2004; Trachtenberg, et al. 2012). We have successfully employed microarray technology in the context of environmental monitoring (Baker, et al. 2009; Baker, et al. 2013)
DNA microarrays, however, rely on the kinetics of probe binding and consequently low abundance transcripts may not be detected owing to poor signal to noise ratios. A pitfall is that many commercial catalog microarrays are frozen in time, in that their design may be based on a draft or early genome build for an organism. It is important, when working with zebrafish microarrays, to verify that current annotation has been used in the array design. Older array designs derived from draft genome builds, when the genome annotation lacked the sophistication and refinement of the current reference sequence, may contain probes that are no longer relevant and also may lack critical probes (Hardiman 2004; Trachtenberg et al. 2012).
Massively parallel sequencing
A major technological shift in the research community in the past five-year period has been the adoption of high throughput sequencing (HTS) technologies to facilitate whole genome and transcriptome sequencing (ten Bosch and Grody 2008; Tucker, et al. 2009). Conventional sequencing approaches were both labor intensive and costly. High molecular DNA fragments were broken down into smaller pieces, which were amplified and sequenced individually using fluorescent dideoxy-nucleotides triphosphates (ddNTPs) as DNA chain terminators. Capillary-based electrophoresis separated these DNA fragments, and facilitated the detection and recording of fluorescence, one base or fragment at a time, yielding short reads 100 to 1000 bp in length, which were assembled into longer contiguous reads. The main advantage of ‘next-generation’ or ‘second-generation’ technologies is that complex DNA cloning and library construction are avoided and shotgun approaches are implemented where DNA is randomly fragmented and many DNA fragments are sequenced simultaneously (Bhasker and Hardiman 2010; Hardiman 2008). A detailed description of the technologies and the inherent biases associated with each of these new technologies is beyond the scope of this review. However we guide the reader to the following articles (Boland, et al. 2013; Carneiro, et al. 2012; Frey, et al. 2014; Glenn 2011; Lam, et al. 2012; Liu, et al. 2012; Meacham, et al. 2011; Minoche, et al. 2011; Pareek, et al. 2011; Quail, et al. 2012; Suzuki, et al. 2011).
To date the majority of the published studies combining RNA-sequencing and zebrafish have focused on immunity (Aday, et al. 2011; Hegedus, et al. 2009; Soares, et al. 2009). Other biological questions are being addressed using high throughput sequencing include development, microRNA discovery and chromatin modification (Aanes, et al. 2011; Aday et al. 2011; Soares et al. 2009). A detailed review of the advantages and applications of these technologies is beyond the scope of this review, but five of the most commonly used platforms are discussed below. The salient features of these key HTS technologies are summarized in Table 1. Applications and the strengths and weaknesses of these platforms are summarized in Table 2.
Table 1.
High Throughput Sequencing Platforms; Chemistry, Read Lengths and Run times
| Platform | Chemistry | Read length (bp) | Per full run | Run time |
|---|---|---|---|---|
| Roche | Pyrosequencing | |||
| GS Junior System | 400 | 35 Mb | 10 h | |
| GS FLX+ System | Up to 1,000 (with GS FLX Titanium XL+) | 700 Mb | 23 h | |
| Illumina | Sequencing by synthesis | |||
| MiSeq | 2 × 300 (MISEQ REAGENT KIT V3) | 13.2–15 Gb | 65 h | |
| Illumina HiSeq2500 | ||||
| Dual Flow Cell | ||||
| HIGH OUTPUT RUN MODE | 2 × 100 | 540–600 Gb | 11 d | |
| RAPID RUN MODE | 2 × 150 | 150–180 Gb | 40 h | |
| ABI SOLiD | Sequencing by ligation | |||
| 5500xl System | Mate-paired: 2 × 60 bp | 10–15 Gb/day | 2– 7d | |
| Paired-end: 75 bp × 35 bp | ||||
| Fragment: 75 bp | ||||
| Ion Torrent | Semiconductor Sequencing Technology | |||
| Ion PGM | 400 bp (Ion 318™ Chip v2) | 600 Mb- 1 Gb | 7.3 h | |
| Ion Proton | Up to 200-base fragment reads (with Ion PI™ Chip) | Up to 10 Gb | 2–4 h | |
| Pacific Biosciences | Single Molecule Real Time | |||
| Pacific Biosciences PacBio RS | 4200 bp mean mapped read length (with XL Binding/C2 Sequencing Chemistry) | 220 Mb | 2 × 55 m (per SMRT cell) | |
| 7600 bp mean mapped read length (with P5/C3 Sequencing Chemistry) | 608 Mb | 180 m (per SMRT cell) |
Table 2.
High Throughput Sequencing Platforms; Applications, Strengths and Weaknesses
| Platform | Advantages | Disadvantages | Applications |
|---|---|---|---|
| Roche | |||
| GS Junior System | Short run time, long reads | Low sequencing depth | Amplicon |
| GS FLX+ System | Sequence Capture | ||
| Whole Genome | |||
| Metagenomics | |||
| Transcriptome | |||
| Illumina | |||
| MISEQ | high throughput | DNA sequence amplification bias | De novo genomes, transcriptomes, metagenomes |
| Illumina HiSeq2500 | |||
| Dual Flow Cell | |||
| HIGH OUTPUT RUN MODE | Ultra-high throughput (Up to 3 billion single reads or 6 billion paired-end reads) | Amplification bias | De novo genomes, transcriptomes, metagenomes |
| RAPID RUN MODE | Ultra-high throughput Up to 600 million single reads or 1.2 billion paired-end reads | Amplification bias | De novo genomes, transcriptomes, metagenomes |
| ABI SOLiD | |||
| 5500xl System | Ultra-high throughput | Amplification bias | De novo genomes, transcriptomes, metagenomes |
| Ion Torrent | |||
| Ion PGM | high throughput | Amplification bias | Targeted sequencing, Copy number analysis, de novo small genome sequencing, |
| Metagenomics, ChIP sequencing, Methylation analysis | |||
| Ion Proton | high throughput (60–80 million reads passing filter) | Amplification bias | Genome, Exome, Gene, ChIP, Methylation, Transcriptome sequencing |
| Pacific Biosciences PacBio RS | Long read sequencing | Low single pass accuracy ~85% | Small Genome Sequencing, Targeted Sequencing, DNA Base Modification Analysis |
454 Sequencing
The first commercial next generation sequencing platform was introduced in 2005 by 454 Life Sciences (Branford, CT, USA) a subsidiary of Roche (Basel, Switzerland). Since then, there has been phenomenal growth and improvement of high throughput sequencing technologies. 454 Sequencing employs a large-scale parallel pyro-sequencing system routinely capable of sequencing approximately 400–600 megabases of DNA per 10-hour run on the Genome Sequencer FLX with GS FLX Titanium series reagents (Voelkerding, et al. 2009). More recent advances with the GS FLX+ System facilitate read lengths up to 1,000 bp in length. 454 sequencing has been used for whole genome sequencing, transcriptome sequencing, amplicon sequencing, capture based sequencing and metagenomics analyses of complex environmental samples (Table 1).
454 sequencing has been used in metagenomic studies where 16S rRNA gene sequencing was performed to investigate EDC-degrading microbial enrichment cultures (Villemur, et al. 2013). 454 sequencing was also used to examine the impact of a known endocrine disruptor (tributyltin or TBT) on the transcriptome of the dog whelk, Nucella lapillus (Chapman and Guillette 2013; Pascoal, et al. 2013). This revealed that TBT mimics the endogenous ligand of the nuclear retinoid X receptor (RXR) and/or peroxisome profilerator-activated receptor (PPAR) disrupting pathways.
454 sequencing was utilized for miRNA discovery in zebrafish. Twenty five novel miRNAs were predicted. Additionally 107 miRNA star sequences and also 41 candidate miRNA targets were identified. A miRNA expression profile built on the basis of pyrosequencing read numbers demonstrated high expression of most miRNAs throughout zebrafish development and identified tissue specific miRNAs (Soares et al. 2009).
To date 454 sequencing technology has not been used for the study of endocrine disruption in zebrafish. The strength of this sequencing approach is the long read data. This permits optimization of de novo transcriptome assembly from next-generation sequencing data in the absence of a scaffold reference in zebrafish strains exposed to endocrine disruptors. This technology has been useful to date in analysis of transcriptomes from the Atlantic cod (Gadus morhua) including RNA transcripts in cells and tissues from various life stages, tissue types, physiological states, and environmental conditions (Johansen, et al. 2011). It has also facilitated de novo assembly of the guppy (Poecilia reticulata) transcriptome (Fraser, et al. 2011).
The competitive landscape of high-throughput sequencing and the disruptive nature of this rapidly evolving technology are highlighted by the fact that that 454 sequencing will soon be obsolete. The platform will cease to be supported by Roche by mid-2016.
Illumina platforms
The HiSeq 2500 (Illumina) employs a proprietary clonal array approach, where individual DNA molecules are fixed to a flat flow-cell surface and are amplified in situ, and serve as templates for synthetic sequencing with novel fluorescent reversible terminator chemistry (Bentley, et al. 2008). With the Illumina approach, isothermal amplification of DNA constructs is carried out creating clonal template clusters (approximately 1,000 copies each). The Illumina HiSeq instruments directly sequence the resulting high-density array of template clusters on the flow cell using sequencing by synthesis (SBS). Four proprietary fluorescently labeled, reversible terminator nucleotides are utilized to sequence the millions of clusters base by base in parallel. The surface is subsequently imaged to generate massive amounts of DNA sequence data (Bentley et al. 2008).
This platform yields high quality short sequence tags (up to 150 bp in length), outputting up to 600 Gb sequence per instrument run, and has enabled high-throughput genome and transcriptome analyses for thousands of organisms. A related instrument the Illumina desktop MiSeq sequencer enables longer sequence tags (up to 300 bp in length) and an output of up to 15 Gb sequence per instrument run.
In early 2014, Illumina presented two new sequencing platforms, the NextSeq 500 and the HiSeq X Ten. The NextSeq 500 represents a novel sequencing-by-synthesis approach based on a two channel approach, single dyes for the bases adenine and cytosine, two dyes for the base thymine, and a dark state for the base guanine. This desktop instrument yields sequence tags (up to 150 bp in length), outputting up to 120 Gb sequence per instrument run. The X Ten has been designed with one focused application, whole human genome sequencing. One instrument run generates 600 Gb of sequence per day which is the equivalent of five human genomes sequenced at 40x genome coverage.
Over 90% of all sequencing data generated to date has been with Illumina sequencers (Hardiman 2008; Voelkerding et al. 2009) and based on continuing innovations in chemistry, optics, flow-cell design and bioinformatics approaches for data analyses this platform will continue to be a major contributor to large scale sequencing projects for the foreseeable future. Additionally the cost per bp of DNA sequence will continue to drop.
This technology has been successful in sequencing human genomes (Levy, et al. 2007). The short individual read lengths have led to primary applications in re-sequencing, where an established reference genome exists rather than de novo sequencing. Cancer genomes are routinely sequenced to great depth using Illumina sequencing. The effects of tobacco smoke on a small-cell lung cancer cell line, NCI-H209 was investigated using Illumina sequencing, providing in depth views of mutational processes, cellular repair pathways and gene networks associated with cancer (Pleasance, et al. 2010). Genome wide location studies have been extensively performed with Illumina sequencing. ChIP-seq combines chromatin immunoprecipitation (ChIP) with HT sequencing to uncover the binding sites of DNA-associated proteins. (Barski, et al. 2007; Johnson, et al. 2007).
Illumina sequencing has been used to investigate the effects and mechanisms of toxicity of silver nanoparticles in zebrafish. In recent years, nanoparticles have been increasingly used in several industrial, consumer and medical applications because of their unique physio-chemical properties. Low concentrations of silver nanoparticles can cause significant disruptions to natural ecosystems (Colman, et al. 2013). The detrimental health effects of nanoparticles remain to be fully elucidated, but there is increasing interest in the role as potential endocrine disruptors (Iavicoli, et al. 2013). The effects of exposures to silver in different forms, (nano, bulk, and ionic forms) was investigated by exposing zebrafish embryos and carrying out transcriptomic analysis using High-Throughput SuperSAGE (van Aerle, et al. 2013). This approach is an advanced form of conventional serial analysis of gene expression technology (SAGE). A specific sequence tag from each transcribed gene is mapped, identified and counted. By sequencing and counting as many tags as possible, a transcriptional profile is provided. Significant alterations in gene expression were uncovered for all forms of silver and many of the gene pathways affected were associated with oxidative phosphorylation and protein synthesis. There was a strong overlap between the different forms of silver. This indicates similar mechanisms of toxicity between the different forms of silver and suggests a requirement for bioavailability of silver ions.
Arsenic is a worldwide and mobile metalloid pollutant in environment. It is highly toxic to most species, although a few species of bacteria utilize arsenic compounds as respiratory metabolites. Arsenic contamination of groundwater is a problem that affects millions of people worldwide (Fendorf, et al. 2010). RNA-SAGE (serial analysis of gene expression) using zebrafish has been used to investigate the molecular mechanism of arsenate toxicity (Xu, et al. 2013). Utilizing 12 million SAGE tags mapped to the zebrafish genome, transcriptional profiles revealed that differentially expressed genes were significantly enriched in several major biological processes including oxidation reduction, translation, iron ion transport, cell redox, and homeostasis. Accordingly, the main pathways disturbed by arsenic treatment include metabolic pathways, proteasome, oxidative phosphorylation, and cancer. Pathway Analysis identified a network with four important hub genes, including Jun, Kras, APoE and Nr2f2.
RNA sequencing was utilized to study the transcriptome of the zebrafish pineal gland and the effects on circadian clockwork by light exposure (Ben-Moshe, et al. 2014). The authors focused on light-induced genes that encode transcription factors. They noted that two factors, dec1, in addition to the core clock gene per2, were critical for the light-entrainment of rhythmic locomotor activity.
Applied Biosystems SOLiD DNA sequencing
Agencourt Personal Genomics introduced ‘supported oligo ligation detection’ (SOLiD) DNA sequencing technology which was subsequently developed and commercialized by Life Technologies (Applied Biosystems/Foster City, CA). It uses a different approach to Illumina ‘sequencing by synthesis’ termed ‘sequencing-by-ligation’. With SOLiD sequencing, a DNA library is prepared and used to generate clonal magnetic bead populations. Each bead contains a unique fragment species. Emulsion PCR generates DNA template which is subsequently covalently attached to a glass surface. Sequencing employs four fluorescent tags and a two-base readout system. Every ligation step interrogates a pair of adjacent nucleotides. As each base is effectively interrogated twice, this results in high accuracy of the sequence calls. Read lengths are mate-paired: 2 × 60 bp; paired-end: 75 bp × 35 bp and fragment: 75 bp. Applications to date have included analyses of sequence variations including, copy number variations (CNVs), single base duplications, inversions, insertions and deletions in addition to SNPS (single nucleotide polymorphisms). A high-resolution, nucleosome position map of the nematode C. elegans was generated using SOLiD. This technology facilitated a global view of the chromatin architecture of a multicellular animal at extremely high density and resolution (Valouev, et al. 2008).
Owing to the different metabolic requirements for male and female reproduction, the liver is one of the most sexually dimorphic organs in terms of its transcriptomes. The liver has been the primary focus of many studies on endocrine disruption as it is the key organ involved in detoxification and responses to estrogens and other hormones. In oviparous species, such as zebrafish, the female liver is the main organ for production of yolk protein precursors (vitellogenins) and some zona pellucida proteins. High throughput sequencing using SAGE was carried out to delineate the transcriptome profiles in male and female zebrafish (Xu et al. 2013). Construction of SAGE libraries and sequencing were performed using SOLiD™ Analyzer 4 (Applied Biosystems). In zebrafish, sexual dimorphism in xenobiotic metabolism and anti-oxidation gene expression was reported. This indicated that retinol x receptor (RXR) and liver x receptor (LXR), important targets for endocrine disruption, play central roles in regulating the sexual differences of lipid and cholesterol metabolisms.
Ion Semiconductor sequencing
Ion Semiconductor Sequencing (Ion Torrent Systems Inc.) represents another platform that has become popular in recent years. Rothberg and colleagues described this post light sequencing approach that was unique in that it was not dependent on conventional fluorescence, complex optics or modified nucleotides. Low-cost semiconductor manufacturing techniques were utilized to generate an integrated circuit capable of direct non-optical DNA sequencing using natural nucleotides. The sequencer contains an electronic reader board which interfaces with the sequencing chip, a signal processing microprocessor, and a fluidics system to control reagents flow over the chip. The ions generated by the DNA polymerase catalyzed synthesis are detected via an ion sensor. Instrument cost is diminished as there are no optical components. Two instruments are currently available, the Ion PGM™ and Ion Proton™ with maximal yields of 600 Mb-1 Gb and 10 Gb respectively.
Ion sequencing has evolved as a rapid and cost-effective tool for developing microsatellite markers for non-model fish species. One recent effort sequenced the genome of an endemic fish species (Schizothorax biddulphi) classified as an extremely endangered freshwater fish in China. Sequencing was performed using the Ion torrent PGM™ instrument to obtain a large number of microsatellites for S. biddulphi (Luo, et al. 2012). Ion sequencing has found use in classifying individual bacterial species that comprising complex, polymicrobial patient specimens and has become a tool that complements classical culture-based and molecular microbiology methods. The sequencing data can be used for accurate genus- or species-level taxonomic assignment of the metagenomic communities (Salipante, et al. 2013).
At this time there are no records in the literature of genomic or transcriptomic zebrafish sequencing using Ion Torrent. This is clearly due to the short read lengths and lower depth compared to other technologies. Another challenge with this sequencing has been distinguishing homopolymer repeats of the same nucleotide. However the technology has undergone much development since its initial description and the increased read lengths of 400 bp per run, and greater chip densities for the Ion PGM™ have improved the technology considerably. The relatively low upfront costs, modest operating costs and rapid sequencing speed will see this technology gain traction in zebrafish transcriptional profiling in response to endocrine disruptors.
Single Molecule Real Time sequencing
Distinguished from the approaches described above is long read Pacific Biosciences Single Molecule Real Time (SMRT™) sequencing, a complementary technology which permits observation of natural DNA synthesis by a DNA polymerase as it occurs. SMRT™ DNA sequencing technology uses millions of 10 nm holes (zeromode waveguides, ZMWs) in a silicon dioxide substrate. Each opening has a zeptoliter volume and contains a single DNA polymerase molecule attached to the surface. Incorporation of fluorescent nucleotides into a single strand of DNA is monitored in these ZMWs using a high numerical aperture lens and single photon sensitive CCD array. The SMRT system generates relatively large numbers (100 million) of long sequence reads with average lengths of 3 kb with some sequences extending up to 16 Kb, coupled with a very fast read time of 8 hours for an entire run (Eid, et al. 2009; Korlach, et al. 2010).
The recent global health efforts to sequence and analyze the genomes of the strains of Vibrio cholerae and E. coli in outbreaks in Haiti and Germany respectively have highlighted the considerable power of SMRT™ sequencing in global medicine (Chin, et al. 2011; Rasko, et al. 2011). The ability to sequence microbial genomes in hours (rather than days) with long read lengths is the emerging characteristic of third-generation DNA sequencing technologies such as the Pacific Biosciences platform. Longer reads facilitate easier and faster genome-assembly efforts and provide greater insights into structural variations.
In addition to providing genomic sequences, the Pacific Biosciences technology is unique amongst all others in that it allows mapping of DNA modifications as an integral part of the sequencing protocol. DNA base modifications are important for understanding of biological processes such as gene expression, DNA damage, and DNA repair. The SMRT Analysis pipeline determines the rate of DNA base incorporation during sequencing and transforms the kinetic information for each nucleotide addition into base calls. This permits the platform to accurately distinguish between modified and native bases (Korlach and Turner 2012). Currently this approach is applicable to only smaller microbial genomes, but advances in capture technologies will open this up to larger genomes, such as the zebrafish. At this time there are no records in the literature of genomic or transcriptomic sequencing of zebrafish using Pacific Biosciences technology. Although Pacific Biosciences generates extremely long reads (up to 16kb), the single-pass error rates are high and have been widely publicized (85% accuracy – 15% error) (Koren, et al. 2012; Zhang, et al. 2012). Coverage at a depth of 20X however guarantees a consensus accuracy of 99.999%, (Q50)(Koren et al. 2012). As a consequence the platform has been used to date exclusively for studies of small genomes.
Recent advances in long insert (~20 kb) DNA library preparation methods and Pacific Biosciences P5-C3 sequencing chemistry yielded the first shotgun human genome sequence dataset comprised entirely of long reads from a human DNA sample, the cell line (CHM1htert). The average read length was >7.6 kb, the longest read was 42.7 kb in size with 54x genome coverage. This long read approach will provide insights in structural genomic variation that are difficult to assess using short read technologies.
All of the sequencing technologies described possess positive attributes. Illumina data, for example, are characterized by depth, high base accuracy, compensating for the relatively low single molecule accuracy of the Pacific Biosciences data (Ribeiro, et al. 2012). Conversely, Pacific Biosciences sequence data are generated without amplification, and can provide coverage in regions underrepresented or absent in Illumina data due to amplification bias introduced during the sample preparation. Additionally Pacific Biosciences sequence reads, although lacking the depth of Illumina sequencing, yield read lengths up to 42kb in length, with a median read length of >7 kb, which complement the shorter 50–250 bp Illumina reads (Ribeiro et al. 2012). Pacific Bioscience sequencing has the potential to facilitate higher-quality de novo assemblies of complex genomes including zebrafish, without reliance on conventional reference-guided assemblies. Future sequencing efforts in zebrafish will utilize a combination of approaches to delineate transcriptomic responses in response to EDC exposure.
TRANSCRIPTOME SEQUENCING
Massively parallel sequencing of the RNA content of a cell, tissue or organism (RNAseq) is an innovative technique to interrogate global RNA expression profiles. The transcriptome reflects cellular activity within a tissue at a given point in time. Genome-wide expression studies, which are not influenced by deductive assumptions, can provide an unbiased approach for investigating the effects of endocrine disruption. RNA-Seq is rapidly becoming the method of choice for detecting and quantifying all the genes expressed in a cell (Maher, et al. 2009; Ozsolak and Milos 2011; Wang, et al. 2009).
The zebrafish represents an attractive model for the study of transcriptome perturbations in response to EDCs, as its reference genome is well characterized (Howe et al. 2013). Zebrafish, by virtue of the conservation of many key transcription factors, kinases and other regulatory proteins in its genome and the human genome, is an excellent proxy for assessing the effects of contaminants in the marine environment and for health risk assessment. Coupling high-throughput transcriptomic approaches to the zebrafish model has the potential to provide a powerful tool for comprehensive toxicology studies to properly dissect the immediate and transgenerational elements of EDC action. Deep transcriptome sequencing can potentially uncover the minimum required dosage of an EDC needed to obtain a phenotype, and uncover the mechanism of action of the EDC. This approach has the potential to reveal what dosage levels of EDC pose a threat to human health. Additionally RNAseq of zebrafish in response to EDCs has the potential to provide potentially critical diagnostic markers and targets for therapeutic intervention.
RNAseq allows short fragments of complementary DNA (cDNA) to be sequenced and mapped onto the latest build of the zebrafish reference genome. Unlike microarray data analysis, RNAseq enables identification of transcription initiation sites (TSSs) and new splicing variants, and it permits precise quantitative determination of exon and splicing isoform expression. Another advantage of RNA-Seq over microarray-based methods of detection is in its ability to identify novel transcripts. With most RNAseq approaches, RNA is extracted and PolyA+ mRNA molecules enriched using poly-T oligo-nucleotide attached magnetic beads. The mRNA is fragmented and converted to double stranded cDNA. Illumina (or other platform) platform specific adaptors are ligated to the DNA and HT sequencing is performed. With Illumina sequencing, short reads of 35 bp – 150 bp in length are generated and subsequently aligned to the reference genome. RNA-Seq provides much greater sensitivity compared to microarray analysis particularly when interrogating low abundance transcripts. Additionally the improved ability to discriminate regions of high sequence identity and the more accurate digital measurements makes RNAseq an attractive proposition.
BIOINFORMATICS CONSIDERATIONS
The bioinformatics step in the analysis of data from massive parallel sequencing is a major task with many pitfalls. A schema for transcriptome sequencing and analysis is outlined in Figure 3. Fortunately, a series of new software packages and bioinformatics pipelines optimized for the computational challenges of short-read sequencing have emerged in the past few years (Pepke, et al. 2009; Robertson, et al. 2010; Trapnell and Salzberg 2009; Wolf 2013). Two of these programs include Maq and Bowtie, which use a computational strategy known as ‘indexing’ to speed up their mapping algorithms. Maq is based on spaced seed indexing where a sequence read is broken into four segments or seeds of equal length (Li, et al. 2008). These seeds are paired and stored in a lookup table. Each read is fragmented and seed pairs of seeds are used to query matching positions in the reference. Bowtie uses the Burrows-Wheeler transform to store a memory-efficient representation of the reference genome (Langmead, et al. 2009).
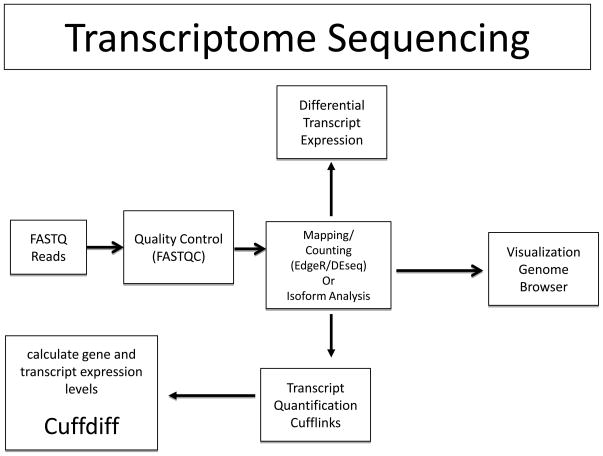
Figure 3.
Schema for transcriptome sequencing and analysis
After the sequence tags have been mapped, the next step typically involves transcript-level quantification. An analytical tool Cufflinks has found widespread use for transcript analysis (Roberts, et al. 2011a; Roberts, et al. 2011b; Trapnell, et al. 2010). Cufflinks does not make use of existing gene annotations during assembly of transcripts, but instead constructs a minimum set of transcripts that best describe the sequence reads present in the dataset. This approach allows Cufflinks to uncover alternative transcription and splicing that are not described by pre-existing gene models. A related program Cuffdiff employs the Cufflinks transcript quantification method and facilitates gene and transcript expression levels across different transcriptomes (Trapnell, et al. 2013). The accuracy of this approach was tested via differential analysis of lung fibroblasts in response to loss of the developmental transcription factor HOXA1, which alters the expression levels of thousands of individual transcripts, in addition to isoform switching events in key cell cycle regulators. Cuffdiff searches for differentially expressed genes by estimating how many fragments derive from each isoform and subsequently converting these counts into isoform expression levels. Cuffdiff thereby facilitates testing for significant differences in isoforms. Cuffdiff relies on a beta negative binomial model to estimate the variance of the RNA-seq data for transcript analysis using t-like statistics from FPKM (fragments per kilobase of exon per million fragments mapped) values.
A different approach to RNA seq analyses utilizes count-based approaches. These methods consider the total output of a locus, without regard to any isoform diversity that may be present. Many tools have been developed for differential expression of read counts, but two of these have gained in popularity, edgeR and DESeq (Anders and Huber 2010; Anders, et al. 2013; Robinson, et al. 2010). The catalyst for development of these tools has been the need has been robust calls of differential transcript expression when working with low numbers of biological replicate samples. Early analysis of RNA employed the Poisson distribution to test for differential expression. The rationale behind this approach was that if sequence reads were independently sampled from a population with given, fixed fractions of genes, the read counts would follow a multinomial distribution, which could be approximated using the Poisson distribution (Marioni, et al. 2008; Wang, et al. 2010). The problem with the application of this approach is that the Poisson distribution is too restrictive, predicting smaller variation that actually existed with the data. Additionally type 1 errors (the probability of false discoveries) were not adequately controlled for. Both EdgeR and DEseq employ a negative binomial distribution which better addresses this issue. A limitation with EdgeR compared to DEseq is that weakly expressed transcripts may be over represented and highly expressed transcripts may not be called as differentially expressed (Anders and Huber 2010).
Platform evaluation for zebrafish transcriptomic analysis
At this point in time a hybrid approach is recommended for the analyses of zebrafish transcriptomes. The sequencing platform should be evaluated on the basis of the goals of the sequencing experiments. If the objective is to understand how environmental pollutants perturb gene expression and alter endocrine signaling pathways, an approach that yields very deep sequencing coverage is recommended (Illumina/Ion Torrent). Biomarkers for pollutant exposures using embryonic, adult male and female zebrafish exposed to different pollutants across a concentration gradient can be performed in parallel. One lane of an Illumina flow cell can generate >200 million reads and this readily permits multiplexing of different samples. The sequencing depth can be optimized to facilitate the analysis of low expressed transcripts. In our experience, 50 million reads permits the analysis of low abundance transcripts. The rationale behind these experiments would be to expose fish to concentrations sufficient to obtain a strong signal for biomarker discovery, guided initially by levels that have been used in the literature to provide sufficient regulation of gene expression and ultimately assessing physiological contaminant levels. If an approach that requires de novo transcriptome assembly or an examination of mRNA variant expression in response to environmental pollutants a mix of Illumina deep sequencing and Pacific Biosciences long read sequencing should be evaluated.
CONCLUSIONS
Ongoing developments promise further advances in the application of RNA seq, particularly direct RNA sequencing and approaches that allow RNA quantification from very small amounts of cellular materials (Maher et al. 2009; Ozsolak and Milos 2011; Wang et al. 2009b). High throughput genome and transcriptome sequencing of zebrafish exposed to EDCs will provide important data sets to better comprehend the risks posed to human health. The zebrafish model is attractive because its short life-span facilitates longitudinal studies. These HTS data sets will provide a better understanding of the number of impacted future generations and their potential for recovery. HTS data sets from embryos and tissues from adults exposed to physiological levels of EDC will provide insights on how developmental mechanisms that integrate genetic and epigenetic interactions are perturbed by EDCs. The development and validation of epigenomic classifiers and biomarkers using zebrafish will provide an important foundation for understanding the molecular basis of EDC toxicity in humans, yielding crucial data on the multigenerational effects of transient exposure during critical stages of development. The data obtained from these studies will permit development of diagnostic and prognostic tests for determining the epigenetic impact of EDCs on human health and for regulating exposure.
Acknowledgments
FUNDING
GH gratefully acknowledges support from NIH grants DK063491, CA023100 and DK080506 and UC Senate grant RK126H-HARDIMAN.
Footnotes
DECLARATION OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the review.
References
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21:1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aday AW, Zhu LJ, Lakshmanan A, Wang J, Lawson ND. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol. 2011;357:450–462. doi: 10.1016/j.ydbio.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Baker M, Šášik R, Gerwick L, Hardiman G. The Praeger Handbook of Environmental Health. 2012. ENDOCRINE DISRUPTORS; pp. 475–502. [Google Scholar]
- Baker ME. Xenobiotics and the evolution of multicellular animals: emergence and diversification of ligand-activated transcription factors. Integrative and Comparative Biology. 2005;45:172–178. doi: 10.1093/icb/45.1.172. [DOI] [PubMed] [Google Scholar]
- Baker ME. Insights from the structure of estrogen receptor into the evolution of estrogens: Implications for endocrine disruption. Biochem Pharmacol. 2011;82:1–8. doi: 10.1016/j.bcp.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Baker ME. What are the physiological estrogens? Steroids. 2013;78:337–340. doi: 10.1016/j.steroids.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Baker ME, Chandsawangbhuwana C. 3D models of MBP, a biologically active metabolite of bisphenol A, in human estrogen receptor alpha and estrogen receptor beta. PLoS One. 2012;7:e46078. doi: 10.1371/journal.pone.0046078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ME, Ruggeri B, Sprague LJ, Eckhardt-Ludka C, Lapira J, Wick I, Soverchia L, Ubaldi M, Polzonetti-Magni AM, Vidal-Dorsch D, et al. Analysis of endocrine disruption in Southern California coastal fish using an aquatic multispecies microarray. Environ Health Perspect. 2009;117:223–230. doi: 10.1289/ehp.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ME, Vidal-Dorsch DE, Ribecco C, Sprague LJ, Angert M, Lekmine N, Ludka C, Martella A, Ricciardelli E, Bay SM, et al. Molecular analysis of endocrine disruption in hornyhead turbot at wastewater outfalls in southern california using a second generation multi-species microarray. PLoS One. 2013;8:e75553. doi: 10.1371/journal.pone.0075553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell JA, McPherson JD, Johnson SL. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Ben-Moshe Z, Alon S, Mracek P, Faigenbloom L, Tovin A, Vatine GD, Eisenberg E, Foulkes NS, Gothilf Y. The light-induced transcriptome of the zebrafish pineal gland reveals complex regulation of the circadian clockwork by light. Nucleic Acids Research. 2014 doi: 10.1093/nar/gkt1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, Belgacem MR, Escriva H. Nuclear hormone receptors in chordates. Mol Cell Endocrinol. 2011;334:67–75. doi: 10.1016/j.mce.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 2004;21:1923–1937. doi: 10.1093/molbev/msh200. [DOI] [PubMed] [Google Scholar]
- Bhasker CR, Hardiman G. Advances in pharmacogenomics technologies. Pharmacogenomics. 2010;11:481–485. doi: 10.2217/pgs.10.10. [DOI] [PubMed] [Google Scholar]
- Boland JF, Chung CC, Roberson D, Mitchell J, Zhang X, Im KM, He J, Chanock SJ, Yeager M, Dean M. The new sequencer on the block: comparison of Life Technology’s Proton sequencer to an Illumina HiSeq for whole-exome sequencing. Hum Genet. 2013;132:1153–1163. doi: 10.1007/s00439-013-1321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SA, et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39:D822–829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003–2004. Environ Health Perspect. 2008;116:39–44. doi: 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, DePristo MA. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics. 2012;13:375. doi: 10.1186/1471-2164-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiz MD, Tso J, Aga DS. Pharmaceutical metabolites in the environment: analytical challenges and ecological risks. Environ Toxicol Chem. 2009;28:2473–2484. doi: 10.1897/09-173.1. [DOI] [PubMed] [Google Scholar]
- Chapman RW, Guillette LJ. Contaminants and impoSEX: transcriptomics of contaminant-induced sex change. Molecular Ecology. 2013;22:1485–1487. doi: 10.1111/mec.12254. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chen H, Hu J, Yang J, Wang Y, Xu H, Jiang Q, Gong Y, Gu Y, Song H. Generation of a fluorescent transgenic zebrafish for detection of environmental estrogens. Aquat Toxicol. 2010;96:53–61. doi: 10.1016/j.aquatox.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Chin C-S, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, et al. The Origin of the Haitian Cholera Outbreak Strain. New England Journal of Medicine. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colborn T, vom Saal FS, Soto AM. Developmental effects of endocrine-disrupting chemicals in wildlife and humans. Environ Health Perspect. 1993;101:378–384. doi: 10.1289/ehp.93101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman BP, Arnaout CL, Anciaux S, Gunsch CK, Hochella MF, Jr, Kim B, Lowry GV, McGill BM, Reinsch BC, Richardson CJ, et al. Low Concentrations of Silver Nanoparticles in Biosolids Cause Adverse Ecosystem Responses under Realistic Field Scenario. PLoS One. 2013;8:e57189. doi: 10.1371/journal.pone.0057189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E, Bourguignon JP, Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT, Gore AC. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocr Rev. 2009;30:293–342. doi: 10.1210/er.2009-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley K, Zon LI. Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Escriva H, Delaunay F, Laudet V. Ligand binding and nuclear receptor evolution. Bioessays. 2000;22:717–727. doi: 10.1002/1521-1878(200008)22:8<717::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Fendorf S, Michael HA, van Geen A. Spatial and temporal variations of groundwater arsenic in South and Southeast Asia. Science. 2010;328:1123–1127. doi: 10.1126/science.1172974. [DOI] [PubMed] [Google Scholar]
- Fraser BA, Weadick CJ, Janowitz I, Rodd FH, Hughes KA. Sequencing and characterization of the guppy (Poecilia reticulata) transcriptome. BMC Genomics. 2011;12:202. doi: 10.1186/1471-2164-12-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey KG, Herrera-Galeano JE, Redden CL, Luu TV, Servetas SL, Mateczun AJ, Mokashi VP, Bishop-Lilly KA. Comparison of three next-generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood. BMC Genomics. 2014;15:96. doi: 10.1186/1471-2164-15-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funder JW. Reconsidering the roles of the mineralocorticoid receptor. Hypertension. 2009;53:286–290. doi: 10.1161/HYPERTENSIONAHA.108.119966. [DOI] [PubMed] [Google Scholar]
- Gao H, Dahlman-Wright K. The gene regulatory networks controlled by estrogens. Mol Cell Endocrinol. 2011;334:83–90. doi: 10.1016/j.mce.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- Gorelick DA, Halpern ME. Visualization of estrogen receptor transcriptional activation in zebrafish. Endocrinology. 2011;152:2690–2703. doi: 10.1210/en.2010-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JD, Clarke CL. Physiological action of progesterone in target tissues. Endocr Rev. 1997;18:502–519. doi: 10.1210/edrv.18.4.0308. [DOI] [PubMed] [Google Scholar]
- Griffitt RJ, Lavelle CM, Kane AS, Denslow ND, Barber DS. Chronic nanoparticulate silver exposure results in tissue accumulation and transcriptomic changes in zebrafish. Aquat Toxicol. 2013;130–131:192–200. doi: 10.1016/j.aquatox.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Grun F, Blumberg B. Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology. 2006;147:S50–55. doi: 10.1210/en.2005-1129. [DOI] [PubMed] [Google Scholar]
- Hardiman G. Microarray platforms--comparisons and contrasts. Pharmacogenomics. 2004;5:487–502. doi: 10.1517/14622416.5.5.487. [DOI] [PubMed] [Google Scholar]
- Hardiman G. Ultra-high-throughput sequencing, microarray-based genomic selection and pharmacogenomics. Pharmacogenomics. 2008;9:5–9. doi: 10.2217/14622416.9.1.5. [DOI] [PubMed] [Google Scholar]
- Hawkins UA, Gomez-Sanchez EP, Gomez-Sanchez CM, Gomez-Sanchez CE. The ubiquitous mineralocorticoid receptor: clinical implications. Curr Hypertens Rep. 2012;14:573–580. doi: 10.1007/s11906-012-0297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus Z, Zakrzewska A, Agoston VC, Ordas A, Racz P, Mink M, Spaink HP, Meijer AH. Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol Immunol. 2009;46:2918–2930. doi: 10.1016/j.molimm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Heindel JJ, vom Saal FS. Role of nutrition and environmental endocrine disrupting chemicals during the perinatal period on the aetiology of obesity. Mol Cell Endocrinol. 2009;304:90–96. doi: 10.1016/j.mce.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, et al. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Henley DV, Korach KS. Endocrine-disrupting chemicals use distinct mechanisms of action to modulate endocrine system function. Endocrinology. 2006;147:S25–32. doi: 10.1210/en.2005-1117. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavicoli I, Fontana L, Leso V, Bergamaschi A. The Effects of Nanomaterials as Endocrine Disruptors. International Journal of Molecular Sciences. 2013;14:16732–16801. doi: 10.3390/ijms140816732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham HA, Redinbo MR. Orphan nuclear receptors adopted by crystallography. Curr Opin Struct Biol. 2005;15:708–715. doi: 10.1016/j.sbi.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Johansen SD, Karlsen BO, Furmanek T, Andreassen M, Jorgensen TE, Bizuayehu TT, Breines R, Emblem A, Kettunen P, Luukko K, et al. RNA deep sequencing of the Atlantic cod transcriptome. Comp Biochem Physiol Part D Genomics Proteomics. 2011;6:18–22. doi: 10.1016/j.cbd.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen BS, Katzenellenbogen JA, Mordecai D. Zearalenones: characterization of the estrogenic potencies and receptor interactions of a series of fungal beta-resorcylic acid lactones. Endocrinology. 1979;105:33–40. doi: 10.1210/endo-105-1-33. [DOI] [PubMed] [Google Scholar]
- Kelce WR, Gray LE, Wilson EM. Antiandrogens as environmental endocrine disruptors. Reprod Fertil Dev. 1998;10:105–111. doi: 10.1071/r98051. [DOI] [PubMed] [Google Scholar]
- Korach KS, Sarver P, Chae K, McLachlan JA, McKinney JD. Estrogen receptor-binding activity of polychlorinated hydroxybiphenyls: conformationally restricted structural probes. Mol Pharmacol. 1988;33:120–126. [PubMed] [Google Scholar]
- Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlach J, Bjornson KP, Chaudhuri BP, Cicero RL, Flusberg BA, Gray JJ, Holden D, Saxena R, Wegener J, Turner SW. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 2010;472:431–455. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- Korlach J, Turner SW. Going beyond five bases in DNA sequencing. Curr Opin Struct Biol. 2012;22:251–261. doi: 10.1016/j.sbi.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Krasowski MD, Ni A, Hagey LR, Ekins S. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR. Mol Cell Endocrinol. 2011;334:39–48. doi: 10.1016/j.mce.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg B, Gustafsson JA. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- Lam HY, Clark MJ, Chen R, Chen R, Natsoulis G, O’Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2012;30:78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambright C, Ostby J, Bobseine K, Wilson V, Hotchkiss AK, Mann PC, Gray LE., Jr Cellular and molecular mechanisms of action of linuron: an antiandrogenic herbicide that produces reproductive malformations in male rats. Toxicol Sci. 2000;56:389–399. doi: 10.1093/toxsci/56.2.389. [DOI] [PubMed] [Google Scholar]
- Lange A, Paull GC, Coe TS, Katsu Y, Urushitani H, Iguchi T, Tyler CR. Sexual reprogramming and estrogenic sensitization in wild fish exposed to ethinylestradiol. Environ Sci Technol. 2009;43:1219–1225. doi: 10.1021/es802661p. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee O, Takesono A, Tada M, Tyler CR, Kudoh T. Biosensor zebrafish provide new insights into potential health effects of environmental estrogens. Environ Health Perspect. 2012;120:990–996. doi: 10.1289/ehp.1104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher RJ, Bustnes JO, Dietz R, Jenssen BM, Jorgensen EH, Sonne C, Verreault J, Vijayan MM, Gabrielsen GW. Exposure and effects assessment of persistent organohalogen contaminants in arctic wildlife and fish. Sci Total Environ. 2010;408:2995–3043. doi: 10.1016/j.scitotenv.2009.10.038. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luccio-Camelo DC, Prins GS. Disruption of androgen receptor signaling in males by environmental chemicals. J Steroid Biochem Mol Biol. 2011;127:74–82. doi: 10.1016/j.jsbmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Nie Z, Zhan F, Wei J, Wang W, Gao Z. Rapid Development of Microsatellite Markers for the Endangered Fish Schizothorax biddulphi (Günther) Using Next Generation Sequencing and Cross-Species Amplification. International Journal of Molecular Sciences. 2012;13:14946–14955. doi: 10.3390/ijms131114946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov GV, Tavares R, Dauphin-Villemant C, Demeneix BA, Baker ME, Laudet V. Independent elaboration of steroid hormone signaling pathways in metazoans. Proc Natl Acad Sci U S A. 2009;106:11913–11918. doi: 10.1073/pnas.0812138106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinerie L, Munier M, Le Menuet D, Meduri G, Viengchareun S, Lombes M. The mineralocorticoid signaling pathway throughout development: expression, regulation and pathophysiological implications. Biochimie. 2013;95:148–157. doi: 10.1016/j.biochi.2012.09.030. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Brain on stress: how the social environment gets under the skin. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17180–17185. doi: 10.1073/pnas.1121254109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham F, Boffelli D, Dhahbi J, Martin DI, Singer M, Pachter L. Identification and correction of systematic error in high-throughput sequence data. BMC Bioinformatics. 2011;12:451. doi: 10.1186/1471-2105-12-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AC, Forbes-Osborne MA, Pillai LS, Fadool JM. Microarray analysis of XOPS-mCFP zebrafish retina identifies genes associated with rod photoreceptor degeneration and regeneration. Invest Ophthalmol Vis Sci. 2011;52:2255–2266. doi: 10.1167/iovs.10-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120474. doi: 10.1098/rstb.2012.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt A, Gumy C. Glucocorticoid and mineralocorticoid action: why should we consider influences by environmental chemicals? Biochem Pharmacol. 2008;76:1184–1193. doi: 10.1016/j.bcp.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Oehlmann J, Oetken M, Schulte-Oehlmann U. A critical evaluation of the environmental risk assessment for plasticizers in the freshwater environment in Europe, with special emphasis on bisphenol A and endocrine disruption. Environ Res. 2008;108:140–149. doi: 10.1016/j.envres.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Oehlmann J, Schulte-Oehlmann U, Kloas W, Jagnytsch O, Lutz I, Kusk KO, Wollenberger L, Santos EM, Paull GC, Van Look KJ, et al. A critical analysis of the biological impacts of plasticizers on wildlife. Philos Trans R Soc Lond B Biol Sci. 2009;364:2047–2062. doi: 10.1098/rstb.2008.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek CS, Smoczynski R, Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet. 2011;52:413–435. doi: 10.1007/s13353-011-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoal S, Carvalho G, Vasieva O, Hughes R, Cossins A, Fang Y, Ashelford K, Olohan L, Barroso C, Mendo S, et al. Transcriptomics and in vivo tests reveal novel mechanisms underlying endocrine disruption in an ecological sentinel, Nucella lapillus. Molecular Ecology. 2013;22:1589–1608. doi: 10.1111/mec.12137. [DOI] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6:S22–32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins GS, Ye SH, Birch L, Ho SM, Kannan K. Serum bisphenol A pharmacokinetics and prostate neoplastic responses following oral and subcutaneous exposures in neonatal Sprague-Dawley rats. Reprod Toxicol. 2011;31:1–9. doi: 10.1016/j.reprotox.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin C-S, Iliopoulos D, et al. Origins of the E. coli Strain Causing an Outbreak of Hemolytic–Uremic Syndrome in Germany. New England Journal of Medicine. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro FJ, Przybylski D, Yin S, Sharpe T, Gnerre S, Abouelleil A, Berlin AM, Montmayeur A, Shea TP, Walker BJ, et al. Finished bacterial genomes from shotgun sequence data. Genome Res. 2012;22:2270–2277. doi: 10.1101/gr.141515.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011a;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011b;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BS. Bisphenol A: an endocrine disruptor with widespread exposure and multiple effects. J Steroid Biochem Mol Biol. 2011;127:27–34. doi: 10.1016/j.jsbmb.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Saili KS, Tilton SC, Waters KM, Tanguay RL. Global gene expression analysis reveals pathway differences between teratogenic and non-teratogenic exposure concentrations of bisphenol A and 17beta-estradiol in embryonic zebrafish. Reprod Toxicol. 2013;38:89–101. doi: 10.1016/j.reprotox.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salipante SJ, Sengupta DJ, Rosenthal C, Costa G, Spangler J, Sims EH, Jacobs MA, Miller SI, Hoogestraat DR, Cookson BT, et al. Rapid 16S rRNA Next-Generation Sequencing of Polymicrobial Clinical Samples for Diagnosis of Complex Bacterial Infections. PLoS One. 2013;8:e65226. doi: 10.1371/journal.pone.0065226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapolsky RM, Romero LM, Munck AU. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocrine reviews. 2000;21:55–89. doi: 10.1210/edrv.21.1.0389. [DOI] [PubMed] [Google Scholar]
- Schier AF. Genomics: Zebrafish earns its stripes. Nature. 2013;496:443–444. doi: 10.1038/nature12094. [DOI] [PubMed] [Google Scholar]
- Segner H. Zebrafish (Danio rerio) as a model organism for investigating endocrine disruption. Comparative biochemistry and physiology Toxicology & pharmacology: CBP. 2009;149:187–195. doi: 10.1016/j.cbpc.2008.10.099. [DOI] [PubMed] [Google Scholar]
- Sharifi N, Auchus RJ. Steroid biosynthesis and prostate cancer. Steroids. 2012;77:719–726. doi: 10.1016/j.steroids.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Sladek FM. What are nuclear receptor ligands? Mol Cell Endocrinol. 2011;334:3–13. doi: 10.1016/j.mce.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. Parturition. N Engl J Med. 2007;356:271–283. doi: 10.1056/NEJMra061360. [DOI] [PubMed] [Google Scholar]
- Soares AR, Pereira PM, Santos B, Egas C, Gomes AC, Arrais J, Oliveira JL, Moura GR, Santos MA. Parallel DNA pyrosequencing unveils new zebrafish microRNAs. BMC Genomics. 2009;10:195. doi: 10.1186/1471-2164-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenschein C, Soto AM. An updated review of environmental estrogen and androgen mimics and antagonists. J Steroid Biochem Mol Biol. 1998;65:143–150. doi: 10.1016/s0960-0760(98)00027-2. [DOI] [PubMed] [Google Scholar]
- Soto AM, Rubin BS, Sonnenschein C. Interpreting endocrine disruption from an integrative biology perspective. Mol Cell Endocrinol. 2009;304:3–7. doi: 10.1016/j.mce.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto AM, Vandenberg LN, Maffini MV, Sonnenschein C. Does breast cancer start in the womb? Basic Clin Pharmacol Toxicol. 2008;102:125–133. doi: 10.1111/j.1742-7843.2007.00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Barros RPA, Warner M, Gustafsson J-A. ERbeta: recent understanding of estrogen signaling. Trends in endocrinology and metabolism: TEM. 2010;21:545–552. doi: 10.1016/j.tem.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Ono N, Furusawa C, Ying BW, Yomo T. Comparison of sequence reads obtained from three next-generation sequencing platforms. PLoS One. 2011;6:e19534. doi: 10.1371/journal.pone.0019534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan SH. Fetal and postnatal environmental exposures and reproductive health effects in the male: recent findings. Fertil Steril. 2008;89:e45. doi: 10.1016/j.fertnstert.2007.12.027. [DOI] [PubMed] [Google Scholar]
- Tamura H, Maness SC, Reischmann K, Dorman DC, Gray LE, Gaido KW. Androgen receptor antagonism by the organophosphate insecticide fenitrothion. Toxicol Sci. 2001;60:56–62. doi: 10.1093/toxsci/60.1.56. [DOI] [PubMed] [Google Scholar]
- ten Bosch JR, Grody WW. Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. J Mol Diagn. 2008;10:484–492. doi: 10.2353/jmoldx.2008.080027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JW, Walker EA, Bujalska IJ, Draper N, Lavery GG, Cooper MS, Hewison M, Stewart PM. 11beta-hydroxysteroid dehydrogenase type 1: a tissue-specific regulator of glucocorticoid response. Endocr Rev. 2004;25:831–866. doi: 10.1210/er.2003-0031. [DOI] [PubMed] [Google Scholar]
- Trachtenberg AJ, Robert JH, Abdalla AE, Fraser A, He SY, Lacy JN, Rivas-Morello C, Truong A, Hardiman G, Ohno-Machado L, et al. A primer on the current state of microarray technologies. Methods Mol Biol. 2012;802:3–17. doi: 10.1007/978-1-61779-400-1_1. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nat Biotechnol. 2009;27:455–457. doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker T, Marra M, Friedman JM. Massively Parallel Sequencing: The Next Big Thing in Genetic Medicine. The American Journal of Human Genetics. 2009;85:142–154. doi: 10.1016/j.ajhg.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda AM, Barber LB, Gray JL, Lopez EM, Woodling JD, Norris DO. Reproductive disruption in fish downstream from an estrogenic wastewater effluent. Environ Sci Technol. 2008;42:3407–3414. doi: 10.1021/es0720661. [DOI] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aerle R, Lange A, Moorhouse A, Paszkiewicz K, Ball K, Johnston BD, de-Bastos E, Booth T, Tyler CR, Santos EM. Molecular mechanisms of toxicity of silver nanoparticles in zebrafish embryos. Environ Sci Technol. 2013;47:8005–8014. doi: 10.1021/es401758d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM. Bisphenol-A and the great divide: a review of controversies in the field of endocrine disruption. Endocr Rev. 2009;30:75–95. doi: 10.1210/er.2008-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemur R, Dos Santos SC, Ouellette J, Juteau P, Lepine F, Deziel E. Biodegradation of endocrine disruptors in solid-liquid two-phase partitioning systems by enrichment cultures. Appl Environ Microbiol. 2013;79:4701–4711. doi: 10.1128/AEM.01239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihua Z, Lathe R, Warner M, Gustafsson J-A. An endocrine pathway in the prostate, ERbeta, AR, 5alpha-androstane-3beta,17beta-diol, and CYP7B1, regulates prostate growth. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13589–13594. doi: 10.1073/pnas.162477299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB. Principles of transcriptome analysis and gene expression quantification: an RNA-seq tutorial. Mol Ecol Resour. 2013;13:559–572. doi: 10.1111/1755-0998.12109. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly Conserved Non-Coding Sequences Are Associated with Vertebrate Development. PLoS Biol. 2004;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Lam SH, Shen Y, Gong Z. Genome-Wide Identification of Molecular Pathways and Biomarkers in Response to Arsenic Exposure in Zebrafish Liver. PLoS One. 2013;8:e68737. doi: 10.1371/journal.pone.0068737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Ishibashi H, Kohra S, Arizono K, Tominaga N. Short-term effects of endocrine-disrupting chemicals on the expression of estrogen-responsive genes in male medaka (Oryzias latipes) Aquat Toxicol. 2005;72:239–249. doi: 10.1016/j.aquatox.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Zhang X, Davenport KW, Gu W, Daligault HE, Munk AC, Tashima H, Reitenga K, Green LD, Han CS. Improving genome assemblies by sequencing PCR products with PacBio. Biotechniques. 2012;53:61–62. doi: 10.2144/0000113891. [DOI] [PubMed] [Google Scholar]
- Zhou J, Cidlowski JA. The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids. 2005;70:407–417. doi: 10.1016/j.steroids.2005.02.006. [DOI] [PubMed] [Google Scholar]