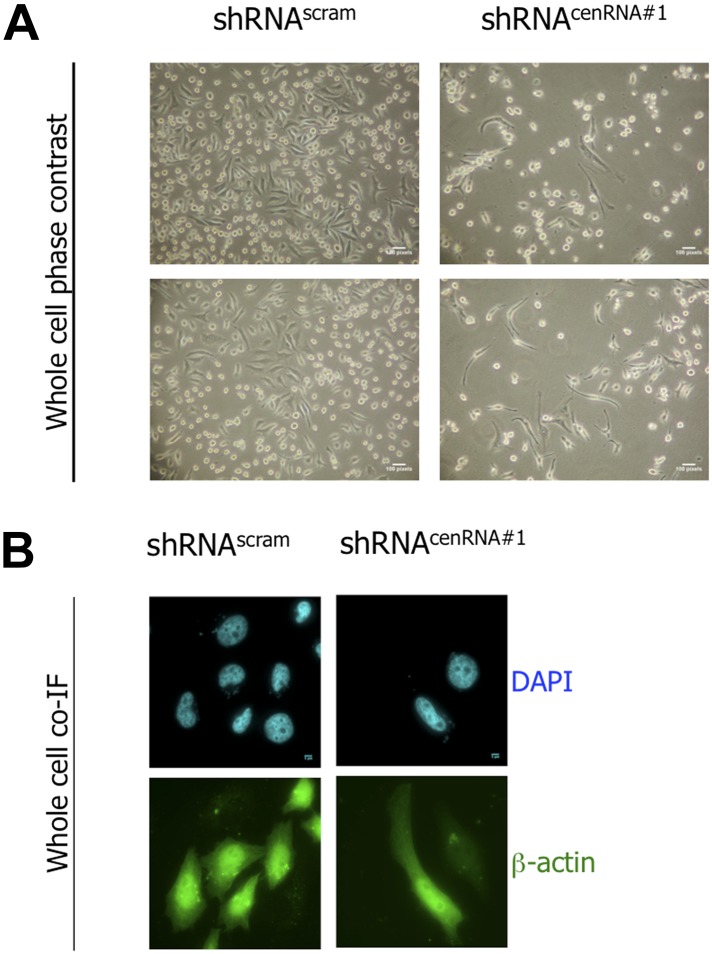
Figure 5. The identified cenRNA#1 transcript associated with CENP-A has a centromeric origin.
(A) CENP-A (green) was co-stained with Xist DNA FISH probe (FISH XistDNA, red) on chromatin fibers (cyan, DAPI). Two independent experiments were performed and in each, a minimum of eight chromatin fibers were analyzed per slide (co-localization on the same chromatin fiber between CENP-A and FISHXistDNA = 0/18). Scale bar: 1 µ m. (B) Chromatin fibers were either co-stained by IF/FISH with CENP-A (green) and cenRNA#1DNA (red), or IF/double-FISH with CENP-A (green), cenRNA#1DNA (red) and centromeric α-satellite probes (gray). The DAPI raw image is shown for a representative chromatin fiber (cyan). Two independent experiments were performed and in each, a minimum of eight chromatin fibers were analyzed per slide (co-localization on the same chromatin fiber between CENP-A and cenRNA#1DNA and FISH α-satellite compared to no co-localization = 9/20 vs 11/20). Scale bar: 1 µm. FISH, fluorescencein situ hybridization; IF, immunofluorescence.