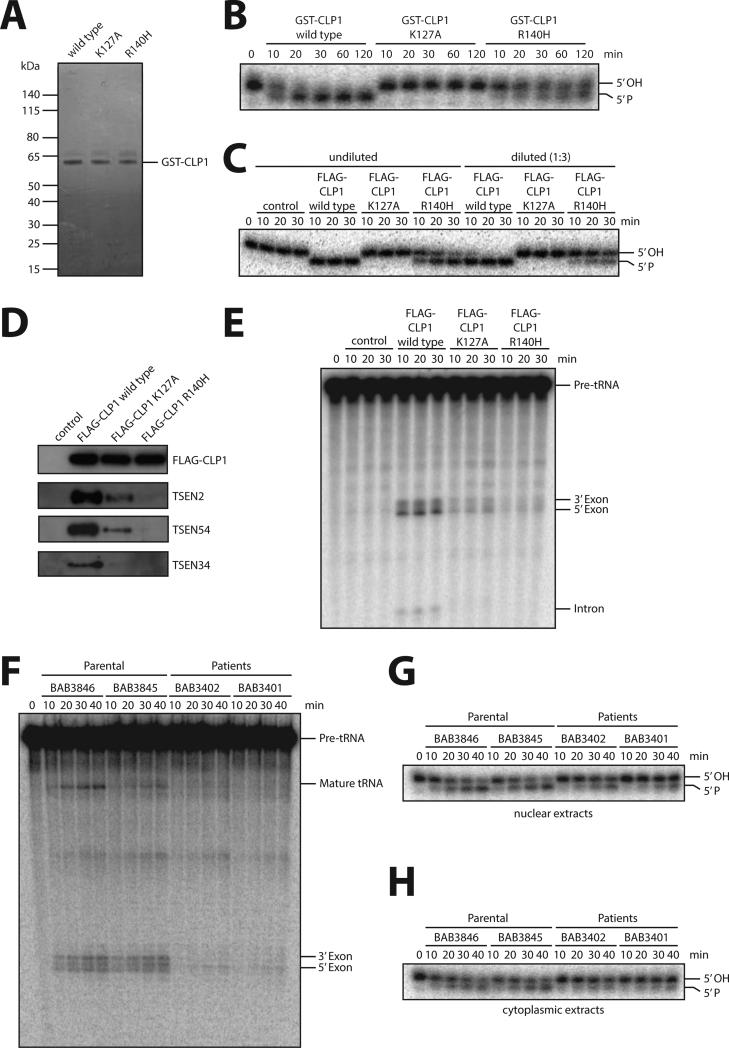
Figure 4. Biochemical studies on purified CLP1 R140H and patient fibroblasts.
(A) Coomassie-blue staining of purified recombinant GST-tagged wild type CLP1, kinase-dead K127A CLP1, and the CLP1 R140H mutant protein. (B) RNA kinase assay using the indicated recombinant CLP1 versions showing that CLP1 R140H is still able to phosphorylate RNA. Recombinant proteins were incubated with an RNA duplex bearing a 5’-OH group and [32P]Cp 3’end-label at one strand for the indicated time points. RNA phosphorylation results in a migration shift after running the reaction products in a denaturing acryamide gel. Note that RNA phosphorylation is completely abolished by the CLP1 K127A mutation. The panel is a representation of two technical replicates. C) RNA kinase assay using protein complexes containing FLAG-CLP1 wild type, FLAG-CLP1 K127A and FLAG-CLP1 R140H affinity-purified from stably expressing HEK293 cells. HEK293 cells without expression of any tagged proteins served as a control. Assays were carried out with undiluted and 1:3-diluted eluates as indicated. (D) Western blotting for TSEN components interacting with affinity-purified FLAG-CLP1 wild type, FLAG-CLP1 K127A and FLAG-CLP1 R140H. (E) Pre-tRNA cleavage assay of affinity-purified FLAG-CLP1 wild type, FLAG-CLP1 K127A and FLAG-CLP1 R140H complexes incubated with an internally labeled intron-containing yeast pre-tRNAPhe. Pre-tRNA processing was monitored by denaturing gel electrophoresis. Panels C-E are representative examples of two replicates. (F) tRNA splicing assay of nuclear extracts of parental (BAB3845 and 3846) and patient (BAB3401 and 3402) fibroblasts incubated for the indicated time points with an internally labeled intron-containing yeast pre-tRNAPhe. Pre-tRNA processing was monitored by denaturing gel electrophoresis. (G, H) RNA kinase activity assay of nuclear (G) or cytoplasmic (H) extracts derived from parental and patient fibroblasts. Extracts were incubated with a 3’ end-labeled 5’ OH group containing RNA duplex for the indicated time points. RNA phosphorylation was monitored by denaturing gel electrophoresis. Panels F-H are representative examples of triplicate experiments. See also Figures S2A to C.