Several decades have passed since the recognition and initial description of the release of extracellular vesicles from cells, but their characterization as potential biomarkers of disease is only now gaining momentum. Recent studies have reported that a variety of extracellular vesicles, such as exosomes and microvesicles, can be isolated from several types of body fluids. The ability to characterize the content and to identify disease-specific molecules such as proteins, mRNA, or non-coding RNA that are expressed on the surface or present within these membrane bound vesicles provides the rationale for their potential value as biomarkers of disease (1, 2). Reports of the identification of extracellular vesicles within bile and their isolation set the stage for analysis of their biomarker potential for biliary tract diseases (3). By examining the microRNA content within extracellular vesicles isolated from bile from three patients with cholangiocarcinoma and three controls, Li et.al. identified a panel of five microRNAs (191, 486-3p, 1274b, 16 and 484) with potential clinical utility as an accurate biomarker for cholangiocarcinoma (4). A sensitivity of 67% and specificity of 96% was reported for diagnosis of a mixed group of 14 patients with intrahepatic, 5 patients with distal, and 27 patients with perihilar cholangiocarcinoma, compared with a heterogeneous group of 50 controls with non-malignant diseases.
The early diagnosis of malignancies that arise from the large biliary ductal system such as perihilar or distal cholangiocarcinoma is difficult (5). When these occur in the setting of biliary tract strictures, it is amongst the most perplexing and challenging aspects of medical practice. Current approaches such as bile duct biopsy or brushings of strictures during endoscopic or radiologic procedures are limited by poor cell yields as a consequence of the desmoplastic nature of the tumor. Attempts to improve the diagnostic yield of cytology, such as by the use of fluorescent in situ hybridization, have had modest impact but continue to be refined. Indeed, existing approaches may be negative in upto 50% of patients with malignancy. By the time more definite signs of cancer arise, tumors may have progressed with spread that precludes curative intervention. Circulating tumor markers such as CA19-9 are not specific and have not been helpful.
An alternative approach has been to examine potential biomarkers within bile. Sampling of bile for the detection of tumor-related biomarkers is appealing for several reasons. First, bile can be obtained at the time of ERCP that may be performed for diagnosis or therapy of biliary tract strictures or suspected biliary tract disease. Next, structuring disease may enhance local concentrations of potential biomarkers that are secreted or released from tumor cells. Concentrations of potential markers may be increased in bile due to impaired bile flow as a result of strictures, or smaller sampling volume of distribution when compared with the systemic circulation. Furthermore, local sampling may have a greater likelihood of detecting candidate biomarkers that are directly related to the tumor. Finally, local sampling may enhance specificity when compared with circulating biomarkers by minimizing the potential for non-specific release from other cells and tissues. Proteomic analysis has identified several biomarkers within bile, including proteins such as CA19-9, Neutrophil gelatinase-associated lipocalin, carcinoembryonic cell adhesion molecule 6, S100 and RNU2-1f (6–8). The diagnostic utility of several of these markers in bile is superior to that of the same markers in serum. Markers such as biliary VEGF-1 may also be useful to exclude cholangiocarcinoma in the distal common bile duct.
Although the study by Li et.al is not the first report of the identification of miRNA within bile (9), or even of their potential as biomarkers, their analysis involved a systematic examination of analysis of miRNA within extracellular vesicles obtained from bile, and resulted in the development of a predictive marker comprised of 5-miRNA that has an improved accuracy for the diagnosis of malignancy within the biliary tract compared with conventional approaches. Their study represents a joint effort involving individuals from several institutions that highlight the advantage and value of such collaborative efforts for ensuring progress in uncommon conditions such as cholangiocarcinoma where the experience in a single-center may be limited. These studies emphasize the utility of bile as a biofluid as well as the analysis of extracellular vesicles for biomarker studies.
There are several challenges and hurdles to be overcome prior to integration of these findings into clinical practice. The effect of bile on either the stability of extracellular vesicles or on the integrity of microRNAs are not known. Bile collection and sampling time with respect to timing of biliary drainage procedures, stability, storage, and the presence of biliary infection are potential confounders that can influence biliary protein concentrations. Whether or not these factors may modulate the release or extracellular vesicles, their specific RNA content or relative RNA concentrations remain to be examined. This is a critical issue for the use of a biomarker assay based on the normalization to input volume as reported by Li. et al. Normalization to endogenous invariant controls would be an optimal choice but data to guide these are lacking. Although small RNA U6 may not be an appropriate choice for analysis of circulating miRNA, we have identified low variance of RNU6B expression within extracellular vesicles that are released from normal as well as malignant hepatocytes and cholangiocytes. In addition to these issues, it is necessary to precisely define the nomenclature and ontology of extracellular vesicles. There exists an extensive literature on the role of microvesicles and microparticles in liver and other diseases. However, these studies encompass a very limited population of extracellular vesicles. The methodologies used for the majority of these studies do not detect all extracellular vesicles, or may include non-vesicular proteins, and may thereby exclude other extracellular vesicles that may have biomarker potential or contribute to functional biological effects. Furthermore, standardization of ontology, as well as of techniques for the identification and characterization of extracellular vesicles are needed. On-going efforts to do so are underway through the auspices of the NIH Common Fund supported Extracellular RNA consortium and other international groups.
Cancer-related miRNA signatures may have clinical utility as markers of disease prognosis. Indeed, circulating miRNAs have been implicated in several biological processes that may be involved and contribute to disease prognosis. Similarly, future studies should be considered to examine the potential for biliary miRNAs as prognostic biomarkers for cholangiocarcinoma. In addition to their biomarker potential, the study of microRNA as well as other non-coding RNA within extracellular vesicles has implications for understanding disease pathobiology. Uptake of extracellular vesicles and release of their biologically active contents by recipient cells provides a mechanism by which cells can interact with each other. Study of intercellular signaling within the local cellular microenvironment has previously been focused on protein mediators such as cytokines and other biologically active factors. The ability of extracellular vesicles to transfer biologically active RNA molecules such as microRNA across cells provides a potent mechanism for intercellular interactions that could be equally as important. Deregulated expression of microRNA in cholangiocarcinoma has now been shown in several studies and justifies further study of the contribution of their transfer via extracellular vesicles on disease pathogenesis and spread. Indeed, extracellular vesicle non-coding RNA content has been reported to vary between malignant and non-malignant hepatocytes. Selected non-coding RNA that are enriched within extracellular vesicles in malignant cells, such as lincROR, have been shown to modulate cellular responses to hypoxia (10). The concept that extracellular vesicle RNA can act as signals to modulate the phenotype within target cells is a novel and emerging paradigm in inter-cellular signaling. This concept has profound implications for understanding the role of intercellular interactions involved in promoting cholangiocarcinoma formation or growth, as well as for other biliary tract diseases. Studies to elucidate the mechanisms by which non-coding RNA within extracellular vesicles are released from tumor cells into bile and their biological relevance are warranted.
Despite several advances in understanding the molecular pathogenesis of cholangiocarcinoma in recent years, their early diagnosis remains an extremely vexing clinical challenge. The release of non-coding RNA within extracellular vesicles into bile may be useful for early diagnosis of malignancy in addition to subserving an important functional biological effect as a mechanism of signaling that mediates cholangiocyte responses to growth stimuli. Further studies to examine other tumor-associated markers such as gene mutations in IDH1 and 2, BAP1 or FGFR2 fusions within extracellular vesicles may allow us to further improve sensitivity for early diagnosis of cholangiocarcinoma.
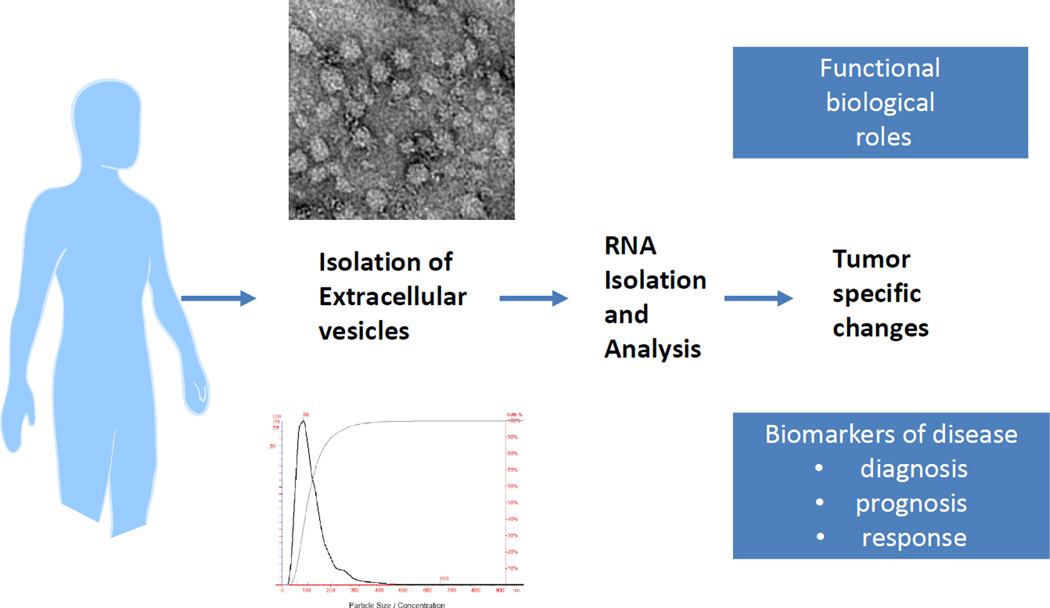
Figure 1.
Extracellular vesicles can be isolated from biological fluids such as bile from patients with cholangiocarcinoma using ultracentrifugation and other techniques. These extracellular vesicles may contain tumor specific proteins or RNA molecules that could provide signatures of the presence of underlying malignancy, and thereby serve as biomarkers of disease, or could have functional biological roles that contribute to the tumoral phenotype.
Acknowledgments
Financial support: Supported in part by Grant DK069370 and TR000884 from the National Institutes of Health
References
- 1.Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer. 2013;4:261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masyuk AI, Huang BQ, Ward CJ, Gradilone SA, Banales JM, Masyuk TV, Radtke B, et al. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. American journal of physiology. Gastrointestinal and liver physiology. 2010;299:G990–G999. doi: 10.1152/ajpgi.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Masica D, Ishida M, Tomuleasa C, Umegaki S, Kalloo AN, Georgiades C, et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014 doi: 10.1002/hep.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel T. Cholangiocarcinoma--controversies and challenges. Nat Rev Gastroenterol Hepatol. 2011;8:189–200. doi: 10.1038/nrgastro.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lankisch TO, Metzger J, Negm AA, Vosskuhl K, Schiffer E, Siwy J, Weismuller TJ, et al. Bile proteomic profiles differentiate cholangiocarcinoma from primary sclerosing cholangitis and choledocholithiasis. Hepatology. 2011;53:875–884. doi: 10.1002/hep.24103. [DOI] [PubMed] [Google Scholar]
- 7.Farina A, Dumonceau JM, Antinori P, Annessi-Ramseyer I, Frossard JL, Hochstrasser DF, Delhaye M, et al. Bile carcinoembryonic cell adhesion molecule 6 (CEAM6) as a biomarker of malignant biliary stenoses. Biochimica et biophysica acta. 2013 doi: 10.1016/j.bbapap.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Alvaro D. Serum and bile biomarkers for cholangiocarcinoma. Current opinion in gastroenterology. 2009;25:279–284. doi: 10.1097/mog.0b013e328325a894. [DOI] [PubMed] [Google Scholar]
- 9.Shigehara K, Yokomuro S, Ishibashi O, Mizuguchi Y, Arima Y, Kawahigashi Y, Kanda T, et al. Real-time PCR-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PloS one. 2011;6:e23584. doi: 10.1371/journal.pone.0023584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular long non-coding RNA regulator of reprogramming. Journal of cell science. 2014 doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]