Abstract
Circulating leukocytes can be used as an effective model to understand the heat stress response of different cattle types and buffaloes. This investigation aimed to determine the temporal profile of HSPs (HSP40, HSP60, HSP70, and HSP90) expression in circulating peripheral blood mononuclear cells (PBMCs) of Murrah buffaloes, Holstein–Friesian (HF), and Sahiwal cows in response to sublethal heat shock at 42 °C. The viability data indicated HF PBMCs to be the most affected to the heat shock, whereas Sahiwal PBMCs were least affected, indicating its better survivability during the heat stress condition. The qRT-PCR expression data showed significant increase in mRNA expression of the analyzed HSPs genes after heat stimuli to the PBMCs under in vitro condition. In each case, the HSPs were most upregulated at 2 h after the heat stress. Among the HSPs, HSP70 was relatively more expressed followed by HSP60 indicating the action of molecular chaperones to stabilize the native conformation of proteins. However, PBMCs from different cattle types and buffaloes showed difference in the extent of transcriptional response. The level of expression of HSPs throughout the time period of heat stress was highest in buffaloes, followed by HF and Sahiwal cows. The higher abundance of HSP70 mRNA at each time point after heat stress showed prolonged effect of heat stress in HF PBMCs. The data presented here provided initial evidence of transcriptional differences in PBMCs of different cattle types and buffaloes and warrant further research.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-013-0486-z) contains supplementary material, which is available to authorized users.
Keywords: Sahiwal (Bos indicus), Murrah Buffalo (Bubalus bubalis), Holstein–Friesian (Bos taurus), PBMCs, Heat stress, Heat shock protein genes, Expression
Introduction
Environmental heat stress negatively affects several dairy parameters including feed intake, milk production, growth, fertility, and conception rates, and it has always been a major concern for livestock industry. The ability of ruminants to regulate body temperature is species and breed dependent. Dairy breeds are typically more sensitive to heat stress than meat breeds, and among dairy breeds, high heat-producing animals are more susceptible as they generate more metabolic heat (Bernabucci et al. 2010). The reduction in milk yield of dairy animals during heat stress (Silanikove et al. 2009) is a major concern for dairy sector world over. Animals can succumb to hyperthermia if they fail to abate the impact of heat stress. The various heat shock proteins (HSPs), that are members of molecular chaperone families, are known to be highly expressed under stressful conditions. These proteins fulfill essential roles under normal conditions and provide protection and adaptation during and following stress (Hartl et al. 2011).
India is home to several zebu cattle (Bos indicus) and riverine buffalo (Bubalus bubalis) breeds that are naturally adapted to different agroclimatic conditions. These breeds are well-known for their hardiness and survival under low input and stressful conditions. The unique thermotolerant ability of Indian native cattle breeds viz. Ongole, Sahiwal, Gir, etc., which outscores the Bos taurus cattle have long been recognized and several synthetic breeds were developed in western countries. Hansen (2004) reviewed the adaptability of zebu and taurine cattle and reported that zebu cattle has better ability to regulate body temperature in response to heat stress than taurine cattle breeds of European origin. Besides native cattle, Indian buffaloes also present an unexplored resource for genetic studies as they portray adaptive trait quite distinct that of cattle breeds. Indian zebu cattle and riverine buffaloes are recognized as major milk producing species in the Indian subcontinent and hence their production performance during summer stress is of great significance to both farmers as well as dairy sector. However, till now no systematic attempts have been made to compare the cellular tolerance of these unique genetic resources towards heat stress. Buffaloes with different skin characteristics and less number of sweat glands suffer from serious problem of heat stress (hyperthermia) than cattle affecting both its reproductive and productive performance at high temperature. There is general agreement in the literature that riverine buffalo are insufficiently heat tolerant and reports indicate that milk yield, growth rate and fertility are all reduced during periods of high ambient temperature (Marai and Haeeb 2010). The differential response of species to heat stress have been documented on the basis of anatomical differences and physiological parameters; however, genetic components, alterations in gene expression and molecular mechanisms underlying changes in heat stress response are not well established in livestock species. Given the complexity of animal physiology and metabolic system, it is difficult to assess the impact of heat stress in dairy animals (Kadzere et al. 2002). However, several attempts have been made in past to understand the negative effect of heat stress in specific cell types (Guerriero and Raynes 1990; Kamwanja et al. 1994; Paula-Lopes et al. 2003; Lacetera et al. 2006; Agnew and Colditz 2008; Patir and Upadhyay 2010; Basirico et al. 2011; Xiang-Hong et al. 2011; Dangi et al. 2012). However, comparative expression profiling of stress responsive genes across different species has not been attempted to date. The expression studies of stress responsive genes to hyperthermia amongst native cattle, exotic cattle and buffaloes would provide comparative baseline data to understand the underlying alterations in cellular tolerance towards heat stress. The present study was planned to assess the comparative changes in gene expression profile of HSP genes in heat stressed peripheral blood mononuclear cells (PBMCs) of Indian cattle (B. indicus), Holstein–Friesian (HF) cattle (B. taurus) and Indian buffaloes (B. bubalis).
Materials and Methods
Sampling
For the present study, a total of 15 lactating animals, 5 each of Murrah buffaloes (B. bubalis), Sahiwal (B. indicus), and HF (B. taurus) cows were included. Sahiwal cows and Murrah buffaloes included in the study belong to cattle farm of National Dairy Research Institute (NDRI), Karnal, while HF cows are maintained at nearby private cattle farm. Blood samples (35–40 mL) were collected in EDTA vacutainers (BD Biosciences) after puncturing the jugular vein of animals. The blood samples were collected at same temperature humidity index of 75.
Isolation of peripheral blood mononuclear cells
For isolating PBMCs, the blood samples were processed within 2 h of sample collection. The whole blood was diluted with 1× PBS (Ca2+ and Mg2+ free; Hyclone, Utah) and was gently over layed on Histopaque-1077 (Sigma-Aldrich Inc., USA). All the steps for PBMCs isolation were performed at room temperature as per manufacturer's instructions. The isolated PBMCs were washed thrice with Ca2+ and Mg2+-free PBS and finally suspended in RPMI 1640 medium (Hyclone, Utah) supplemented with 10 % FCS (PAA). The cells were stained with trypan blue dye and counted using haemocytometer.
Heat stress treatment and cell viability measurement
For each animal, a total of 1 × 106 cells/mL were divided into nine equal aliquots, one aliquot was labeled as basal, four as control (CTR), and four as heat treated (TRT). Initially, all aliquots were incubated at 37 °C for 30 min for stabilization in nutrient media. The aliquots marked as TRT were exposed to heat stress at 42 °C for 1 h. After completion of the heat stress period, the aliquots were allowed to recover at 37 °C. Subsequently, the cells were harvested at different time points (2, 4, 6, and 12 h) post-heat stress. Simultaneous to TRT samples, the CTR (unstressed) samples were also harvested at similar time points for comparison. On the other hand, the basal sample representing the zero time point was harvested after completion of 30 min in stabilization phase. After completion of each time point, small aliquots of cells were used for viability measurement by trypan blue dye exclusion method. The remaining cells were trizolated in ice-cold Trizol (Invitrogen, Carlsbad, California) for RNA extraction.
Total RNA extraction and cDNA synthesis
Total RNA extracted using Trizol was subjected to column purification and RNAse free DNase digestion using RNeasy Mini Kit (Qiagen, Germany). RNA concentration and purity was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) and stored at −80 °C till further usage. cDNA synthesis was performed using 100 ng RNA, 1 μL dT12-18 (Invitrogen Corp. CA), 1 μL 10 mmol/L dNTP mix (Invitrogen Corp., CA), 1 μL random primers (Invitrogen Corp., CA), and 10 μL DNase/RNase free water. The mixture was incubated at 65 °C for 5 min and kept on ice for 3 min. A total of 6 μL of master mix consisting of 4.5 μL 5× first-strand buffer, 1 μL 0.1 M DTT, 0.25 μL (50 U) of SuperScript™ III RT (Invitrogen Corp., CA), and 0.25 μL of RNase Inhibitor (10 U; Promega, WI) was added. The reaction was performed in an Eppendorf gradient cycler using the program: 25 °C for 5 min, 50 °C for 60 min, and 70 °C for 15 min. cDNA was then diluted 1:4 (v/v) with DNase/RNase free water.
Primer designing and quantitative real-time PCR
For qPCR, the primers were either designed using Primer express 3.0 software (Applied Biosystem) or selected from literature. Primer details for the analyzed heat shock protein (HSP) genes are given in Table 1. To check the sequence specificity, primers were aligned using BLASTN at NCBI and UCSC's Cow (B. taurus) genome browser gateway. Prior to qPCR, primers specificity was further confirmed in a 20 μL PCR reaction using the same protocol described for qPCR except for the final dissociation protocol. A 5 μL of the PCR product was analyzed on ethidium bromide stained 2 % agarose gel. The accuracy of primer pairs was also ensured by the presence of a unique peak during the dissociation step at the end of qPCR.
Table 1.
Selected candidate reference genes, primers and different parameters derived from qRT-PCR analysis
| Gene | Description | Biological function | GenBank accession no. | Primers 5′-3′ (forward, reverse) | T m (°C) | Amplicon size (bp) | PCR efficiency (%)a | R 2** |
|---|---|---|---|---|---|---|---|---|
| Internal control genes | ||||||||
| RPS9 1 | Ribosomal protein S9 | Protein synthesis/40S subribosome | DT860044 | CCTCGACCAAGAGCTGAAG and CCTCCAGACCTCACGTTTGTTC | 82.56 | 54 | 86.48 | 0.999 |
| RPS15A | Ribosomal protein S15a | Protein synthesis/40S subribosome | BC108231 | GAATGGTGCGCATGAATGTC and GACTTTGGAGCACGGCCTAA | 82.89 | 101 | 102.87 | 0.997 |
| B2M | Beta-2 microglobulin | Cytoskeletal protein, immune response, protein binding | NM_173893 | CTGCTATGTGTATGGGTTCC and GGAGTGAACTCAGCGTG | 82.39 | 101 | 99.71 | 0.999 |
| Heat shock protein (HSP) genes | ||||||||
| HSP40 | Heat shock protein 40 | Protect proteins from aggregation and cellular stress | NM_001033763.1 | AGCCAGGATCAGCCTTC and AACACAACGGGTATGGT | 84.50 | 84 | 106.00 | 0.973 |
| HSP60 | Heat shock protein 60 | Transportation and refolding of proteins | NM_001166610.1 | CGACAACTTCTGCTGTTGTTA and ATGATGCTATGCTTGGAGAT | 80.29 | 109 | 105.54 | 0.997 |
| HS970 | Heat shock protein 70 | Protein folding, cytoprotection, and as molecular chaperones. | JN604432.1 | AACATGAAGAGCGCCGTGGAGG and GTTACACACCTGCTCCAGCTCC | 86.46 | 171 | 104.71 | 0.991 |
| HSP90 | Heat shock protein 90 | Protein translocation and regulation of steroid hormone receptors | AB072368.1 | CTGTCATCAGCAGTGGG and ACATGCCAACAGGATCTAC | 80.01 | 74 | 90.42 | 0.974 |
Bionaz and Loor (2007)
aqPCR efficiencies for each primer pair was calculated from 6-point standard curves using 5-fold dilution series of pooled cDNA from control and heat-stressed samples
**p <0.01
qPCR was performed using Light Cycler 480 instrument (Roche, Germany) for genes expression study. For each of the studied animal, a total of 9 cDNA samples (one basal; four CTR (2, 4, 6, and 12 h); and four TRT (2, 4, 6, and 12 h)) were analyzed using qPCR. Each reaction in a 96 well white plate (Roche, Germany) was comprised of 4 μL diluted cDNA combined with 6 μL of a mixture composed of 5 μL 2× LightCylcer 480 SYBR Green I master mix (Roche, Germany), 0.4 μL each of 10 μM forward and reverse primers, and 0.2 μL DNase/RNase free water. For each gene, samples were run in duplicate (technical replicates) along with 6 point relative standard curve plus the nontemplate CTR. The reactions were performed with amplification conditions: 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C (denaturation), and 1 min at 60 °C (annealing + extension). A dissociation protocol with an incremental temperature of 95 °C for 15 s plus 65 °C for 15 s was used to investigate the specificity of the qPCR reaction and the presence of primer dimers. The qPCR expression data for each target gene was extracted in the form of crossing point (Cp) values. The data was acquired using the “second derivative maximum” method as computed by the LightCycler Software 3.5 (Roche Diagnostics) and subjected for subsequent analysis.
Normalization and data analysis
For normalization of expression data, three appropriate internal control genes (ICGs) identified in one of our previous study (Kishore et al. 2013) were used. Kishore et al. (2013) evaluated the expression stability of 11 commonly reported ICGs in heat stressed PBMCs of cattle and buffaloes based on stability measures. The analysis identified Beta-2 microglobulin (B2M) and ribosomal proteins (RPS9 and RPS15) as most stably expressed genes and were thus used for normalization of expression data of major HSP genes. The analysis of mRNA expression data across different samples was based on cycle threshold (CT) values. The CT values of each of the target gene were subtracted from the arithmetic mean of CT values of the three ICGs to calculate ΔCT. Results were expressed as means ± SEM. Analysis of variance was performed to determine the presence or absence of significant differences in the analytical variables among different groups using GraphPad PRISM version 5.0 (La Jolla, CA, USA) statistical software package and the differences between groups were tested by Tukey's multiple comparison test. p values less than 0.05 were considered significant.
Results
In this study, PBMCs of Indian Sahiwal cows, HF cows and Murrah buffaloes was utilized as cellular in vitro model to describe the comparative cellular tolerance during heat stress. The cell viability and transcriptional induction of HSP40, HSP60, HSP70, and HSP90 mRNA were used as indicators to evaluate the comparative cellular tolerance ability of different cattle types and riverine buffaloes. The cell viability count for TRT samples was lower than their corresponding CTR (unstressed) throughout the time points under study (Fig. 1 (I–III)). The percent reduction in cell viability especially after 12 h post-heat stress was highest in PBMCs of HF cows followed by Murrah buffaloes and Sahiwal cows. Good quality RNA as reflected A260/A280 ratio of 2.00 ± 0.120 was obtained in all the samples types viz; basal, CTR and heat stressed PBMCs. The melt curve analysis showed single melting peak for each of the analyzed genes indicating quality of qPCR data. Furthermore, high PCR efficiency that ranged from 86.48 to 105.54 (Table 1) was observed different studied genes. In order to normalize the target gene expression data, geometric mean of B2M, RPS9, and RPS15a was considered as ICGs. These three genes were selected as most appropriate for normalization of heat stressed PBMCs in one of our earlier studies (Kishore et al. 2013). In our study, all the PBMCs that were exposed to the elevated temperature of 42 °C were found to be responsive to in vitro heat stress condition. When we examined the time course expression pattern, all the HSP transcripts were upregulated immediately after heat stress in comparison to untreated/CTR cells and remained elevated upto 6 h of recovery phase. Strikingly, the maximum induction in HSP transcripts was observed at 2 h time point post stress and its level was significantly higher (p < 0.05) in comparison to other time points (Fig. 2). At later time points (>6 h post-heat stress), the HSP transcripts declined to a similar level close to CTR/unstressed samples.
Fig. 1.
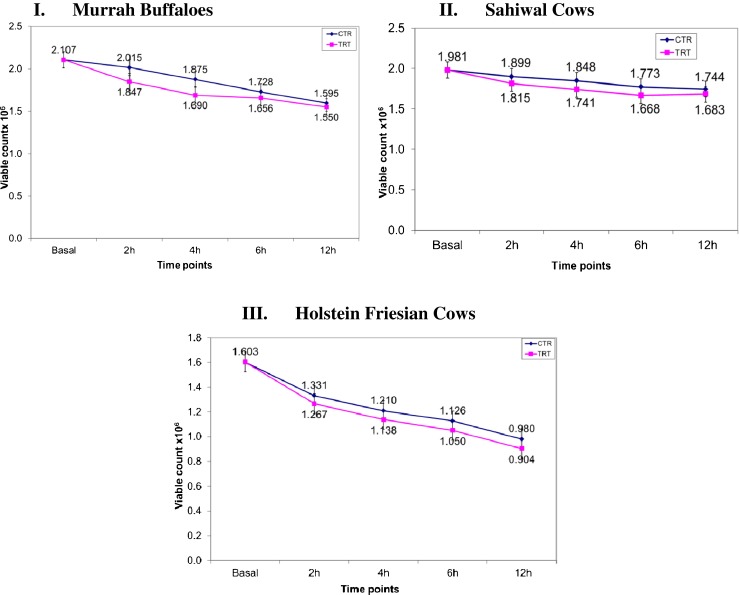
Comparative temporal changes in cellular viability of PBMCs across I Murrah buffaloes, II Sahiwal cows, and III HF cows during in vitro heat stress treatment. Abbreviations: CTR control, TRT heat treated/stressed
Fig. 2.
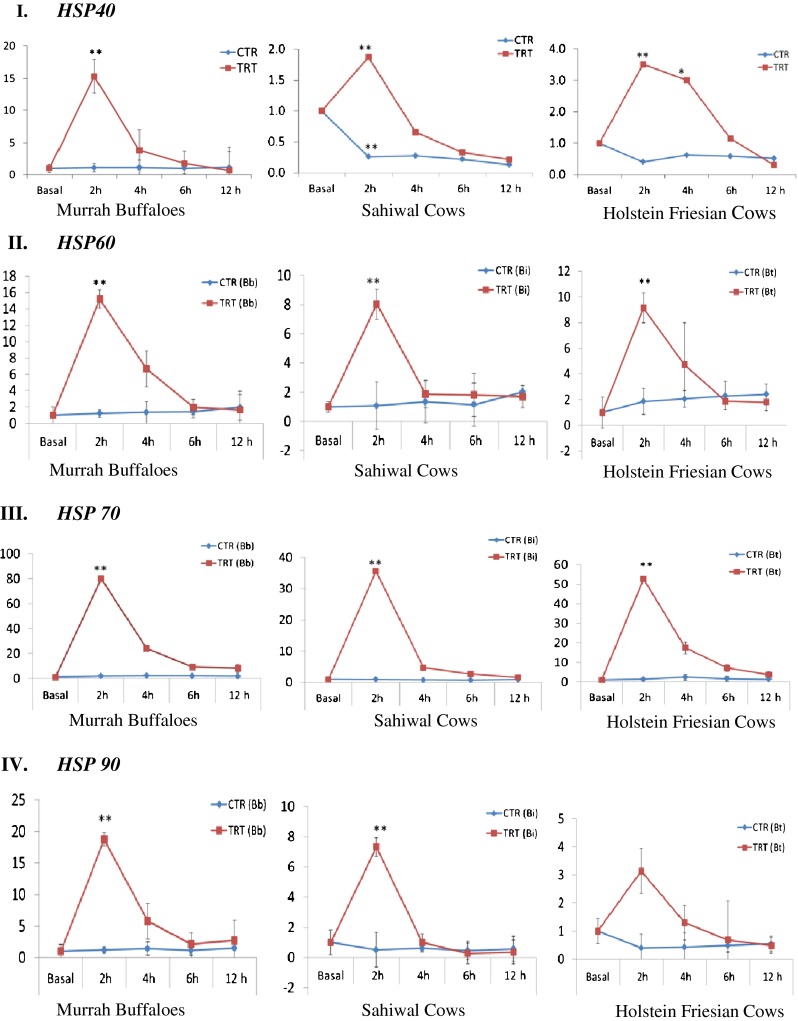
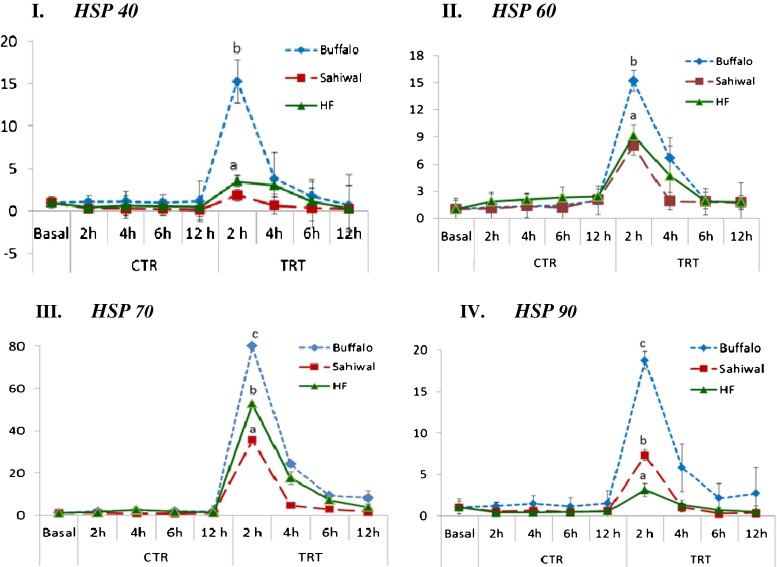
Assesment of relative mRNA expression of individual heat shock protein (HSP) genes in heat stressed (TRT) and unstressed (CTR) PBMCs of Murrah buffaloes, Sahiwal cows, and HF cows. I HSP40, II HSP60, III HSP70, and IV HSP90. For all graphs, y-axis represent relative mRNA expression and x-axis represent the time points. Significant differences (p ≤ 0.05); *p < 0.05 and **p < 0.01. CTR control, TRT heat treated/stressed, Bb B. bubalis, Bi B. indicus, Bt B. taurus, h hour
Under heat shock condition, HSP40 mRNA expression was significantly (p < 0.05) induced at 2 h post-heat stress in PBMCs of Sahiwal cows, HF cows and Murrah buffaloes. The HSP40 transcript was induced maximally at 2 h time point in all the PBMCs (Fig. 2 (I)). Its expression was maximum in Murrah buffaloes, followed by HF and Sahiwal cows (15.27-, 3.5-, and 1.87-fold induction, respectively) (Fig. 3 (I)). HSP60 transcript also showed maximum induction in its expression at 2 h post heat stress (Fig. 2 (II)), with highest induction in Murrah buffaloes, followed by HF and Sahiwal cows (15.2-, 9.14-, 8.04-fold induction, respectively) (Fig. 3 (II)). HSP70 mRNA also showed induction in its expression at 2 h post heat stress (Fig. 2 (III)). At this time point, its expression was highest in Murrah buffaloes (80.0-fold), followed by HF (52.68-fold) and Sahiwal cows (35.64-fold) (Fig. 3 (III)). Like other HSPs, HSP 90 mRNA was also induced maximally at 2 h time point (Fig. 2 (IV)) with highest expression in Murrah buffaloes, followed by Sahiwal and HF cows with expression fold of 18.75, 7.32, and 3.14, respectively (Fig. 3 (IV)).
Fig. 3.
Overall mRNA abundance of different heat shock protein (HSP) genes in heat stressed (TRT) and unstressed (CTR) PBMCs across Murrah buffaloes, Sahiwal cows, and HF cows. I HSP40, II HSP60, III HSP70, and IV HSP90. For all graphs, y-axis represent relative mRNA expression and x-axis represent the time points. Values with different letters are significantly different (p ≤ 0.05); HF Holstein–Friesian, h hour
Additionally, overall induction of different HSP mRNA expression due to heat stress was compared within different animal types and the comparative data is plotted in Fig. 4 (I). In Murrah buffalo PBMCs, HSP70 expression was most pronounced at 2 h post heat stress (fold change, 80.00) followed by HSP90 (fold change, 18.75), HSP40 (fold change, 15.27), and HSP60 (fold change, 15.20). The ranking of genes from higher to lower abundance in Murrah buffaloes were in the following order; HSP70 > HSP90 > HSP40 > HSP60. Similarly, in Sahiwal cows, HSP70 (35.64) was most responsive followed by HSP60 (8.04), HSP90 (7.32), and HSP40 (1.87). The ranking of genes from higher to lower abundance in Sahiwal cows were in the following order; HSP70 > HSP60 > HSP90 > HSP40. In HF cows, HSP70 was observed to be predominantly up regulated (52.68), followed by HSP60 (9.14), HSP40 (3.50), and HSP90 (3.14). The ranking of genes from higher to lower abundance in HF cows were in the following order; HSP70 > HSP60 > HSP40 > HSP90. Our overall data indicated HSP70 chaperon to be most responsive at 2 h post-heat stress in all the animal types included in the study while other HSPs showed moderate to low induction in their expression. Therefore, measuring HSP70 expression level could be utilized as sensitive biomarker to determine the impact of environmental stress in dairy animals.
Fig. 4.
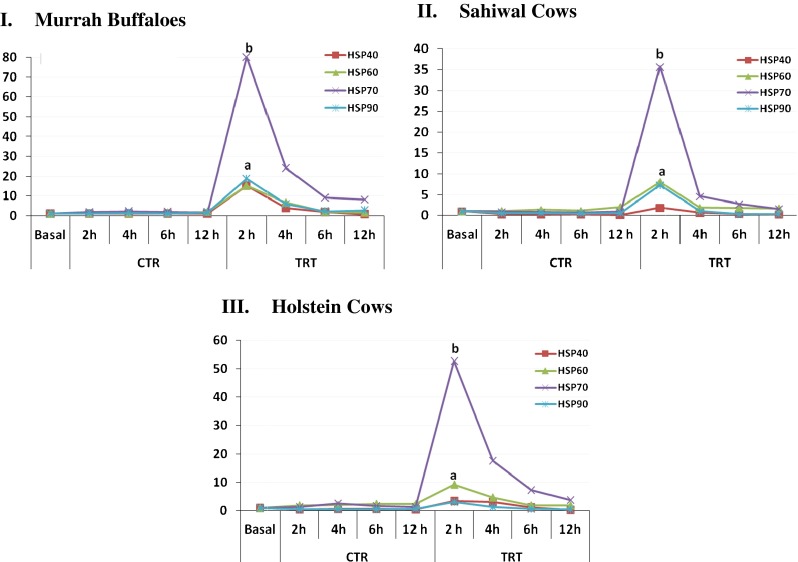
Comparative expression pattern of heat shock protein (HSP) genes in heat stressed (TRT) and unstressed (CTR) PBMCs of I Murrah buffaloes, II Sahiwal, and III HF cows. Abbreviations: CTR control, TRT heat treated/stressed, h hour. Values with different letters are significantly different (p ≤ 0.05)
Discussion
Heat stress affects both physiology and productivity of dairy animals. The duration, intensity and ability to recover from the affect of climatic stress are important factors affecting performance ability of farm animal (Khalifa 2003). The ability of animal to acclimatize and produce under the specific climate condition signifies the adaptation to a particular environmental niche. However, it is not completely understood how dairy animals respond to heat stress and why there is variability at breed or species level to tolerate the environmental heat load. In recent times, much importance has been given to understand the mechanism of molecular adaptation in livestock breeds towards climatic changes especially heat stress by various research groups.
The present study was undertaken with the presumption that circulating PBMCs could serve as suitable in vitro model to understand the heat stress response across different dairy animals. At present, it is not completely understood how dairy animals respond to heat stress and why there is variability at breed or species level to tolerate the environmental heat load. The utility of PBMCs as cellular model to understand stress response in livestock is attributed to its amenability to culturing and responsiveness to heat stress (Guerriero and Raynes 1990; Elvinger et al. 1991; Malayer et al. 1990; Kamwanja et al. 1994; Lacetera et al. 2002).
Several in vitro studies in bovine have shown reduction in cell viability and responsiveness of lymphocytes to mitogens during short and severe heat shock challenge (Elvinger et al. 1991; Kamwanja et al. 1994); Lacetera et al. 2002). However, no comparative study for cattle types and buffalo has been made to understand the cellular response to thermal stress.
The cell viability data in the present study provided clear evidence that PBMCs of Sahiwal cows (B. indicus) which is a naturally adapted breed to tropical environmental conditions of India has better survivability to thermal shock in comparison to Hostein Friesian cows (B. taurus) and buffaloes (B. bubalis). These results are consistent with earlier findings where lesser deleterious effect of high temperature was observed on preimplatation embryos and cells from B. indicus cattle (Malayer and Hansen 1990; Kamwanja et al. 1994; Paula-Lopes et al. 2003; Hansen 2004; Hernandez-Ceron et al. 2004).
Several studies have indicated that constitutive elevation of the inducible HSP level provides cytoprotection upon thermal stress (Horowitz 2001; Sonna et al. 2002; Collier et al. 2008). In this study, expression profile of different HSP transcripts was measured in PBMCs during heat shock condition. The reason for selecting four major HSP genes (HSP70, HSP90, HSP60, and HSP40) for the present analysis was due to their primary role as molecular chaperons that ensures the correct protein folding and apoptosis regulation during physiological stressful conditions. It is also noteworthy that though heat shock response is an evolutionary conserved mechanism, different breeds/species may vary in their inherent ability to induce HSP synthesis during heat shock.
Among genes examined in this study, HSP70 was found to be most dominant transcript expressed in all sample types post heat stress. HSP70 is known to be a highly inducible chaperon with major role to stabilize the native conformation of proteins and maintenance of cell survivability during thermal stress (Beckham et al. 2004). Its expression data is in accordance with studies carried out in different livestock species (Guerriero and Raynes 1990; Kamwanja et al. 1994; Paula-Lopes et al. 2003; Lacetera et al. 2005; Lacetera et al. 2006; Lacetera et al. 2009; Agnew and Colditz 2008; Patir and Upadhyay 2010; Basirico et al. 2011). The difference in extent of HSP70 mRNA expression in PBMCs of Sahiwal, HF cows and buffaloes showed difference in their ability to withstand the deleterious effect of heat stress. The lower expression for HSP70 in Sahiwal cattle provided an evidence for its better cellular tolerance than that of Holstein and buffaloes. This differential expression of HSP70 mRNA in the present study could be attributed to its genetic divergence across the studied species (Sodhi et al. 2013).
In addition to HSP70, other HSP genes that were studied also contribute to thermotolerance such as HSP40, HSP60, and HSP90 (Duncan 2005; Kampinga et al. 2003). It should be noted that HSP40 and HSP90 generally function as a co-chaperone/cofactor of mammalian HSP70 and the complex is involved in the restoring of protein confirmation after heat shock (Welch 1990; Morimoto 1998; Pratt and Toft 2003). HSP40 and HSP90 mRNA expression like other HSPs was highest at 2 h post-heat stress in Murrah. HSP60 is also known to be one of the most important molecular chaperons under various stressful conditions (Oksala et al. 2006) showed higher induction in Murrah buffaloes, followed by HF and Sahiwal cows. The induction of various HSPs reflected their conserved functioning towards protective mechanism against cellular heat stress. The data presented here provides initial evidence of transcriptional differences in PBMCs of different cattle types and warrant further research. Nevertheless, the conclusion can be drawn that: (1) PBMCs as a potential model to understand the differential heat shock response in cattle and buffaloes, (2) HSP70 as most responsive gene and could be utilized as marker for determination of extent of heat stress, and (3) Sahiwal cattle have better cellular tolerance than HF cattle and buffaloes.
Electronic supplementary material
(DOC 39 kb)
Acknowledgments
The work was partly supported by Indian Council of Agriculture Research, New Delhi under National Fellow Scheme. The authors duly acknowledge, Director NBAGR, Karnal for extending the research facilities for this study. The authors also acknowledge Sh Ram Singh, Cattle Farm Taroari, Karnal for providing the HF samples.
References
- Agnew LL, Colditz IG. Development of a method of measuring cellular stress in cattle and sheep. Vet Immunol Immunopathol. 2008;123(3–4):197–204. doi: 10.1016/j.vetimm.2008.01.038. [DOI] [PubMed] [Google Scholar]
- Basirico L, Morera P, Primi V, Lacetera N, Nardone A, Bernabucci U. Cellular thermotolerance is associated with heat shock protein 70.1 genetic polymorphisms in Holstein lactating cows. Cell Stress Chaperon. 2011;16(4):441–448. doi: 10.1007/s12192-011-0257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham JT, Mackanos MA, Crooke C, Takahashi T, O'Connell-Rodwell C, Contag CH, Jansen ED. Assessment of cellular response to thermal laser injury through bioluminescence imaging of heat shock protein 70. Photochem Photobiol. 2004;79:76–85. doi: 10.1111/j.1751-1097.2004.tb09860.x. [DOI] [PubMed] [Google Scholar]
- Bernabucci U, Lacetera N, Baumgard LH, Rhoads RP, Ronchi B, Nardone A. Metabolic and hormonal acclimation to heat stress in domesticated ruminants. Animal. 2010;4(7):1167–1183. doi: 10.1017/S175173111000090X. [DOI] [PubMed] [Google Scholar]
- Bionaz M, Loor JJ (2007) Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol Genomics 29:312–319 [DOI] [PubMed]
- Collier RJ, Collier JL, Rhoads RP, Baumgard LH. Genes involved in the bovine heat stress response. J Dairy Sci. 2008;91:445–454. doi: 10.3168/jds.2007-0540. [DOI] [PubMed] [Google Scholar]
- Dangi SS, Gupta M, Maurya D, Yadav VP, Panda RP, Singh G, Mohan NH, Bhure SK, Das BC, Bag S, Mahapatra R, Sharma GT, Sarkar M. Expression profile of HSP genes during different seasons in goats (Capra hircus) Trop Anim Health Pro. 2012;44(8):1905–1912. doi: 10.1007/s11250-012-0155-8. [DOI] [PubMed] [Google Scholar]
- Duncan R. Inhibition of Hsp90 function delays and impairs recovery from heat shock. FEBS J. 2005;272:5244–5256. doi: 10.1111/j.1742-4658.2005.04921.x. [DOI] [PubMed] [Google Scholar]
- Elvinger F, Hansen PJ, Natzke RP. Modulation of function of bovine polymorphonuclear leukocytes and lymphocytes by high temperature in vitro and in vivo. Am J Vet Res. 1991;52(10):1692–1698. [PubMed] [Google Scholar]
- Guerriero V, Jr, Raynes DA. Synthesis of heat stress proteins in lymphocytes from livestock. J Anim Sci. 1990;68(9):2779–2783. doi: 10.2527/1990.6892779x. [DOI] [PubMed] [Google Scholar]
- Hansen PJ. Physiological and cellular adaptations of zebu cattle to thermal stress. Anim Reprod Sci. 2004;82–83:349–360. doi: 10.1016/j.anireprosci.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hernandez-Ceron J, Chase CC, Hansen PJ. Differences in heat tolerance between preimplantation embryos from Brahman, Romosinuano, and Angus breeds. J Dairy Sci. 2004;87(1):53–58. doi: 10.3168/jds.S0022-0302(04)73141-0. [DOI] [PubMed] [Google Scholar]
- Horowitz M. Heat acclimation: phenotypic plasticity and cues to the underlying molecular mechanisms. J Therm Biol. 2001;26:357–363. doi: 10.1016/S0306-4565(01)00044-4. [DOI] [Google Scholar]
- Kadzere CT, Murphy MR, Silanikove N, Maltz E. Heat stress in lactating dairy cows: a review. Livest Prod Sci. 2002;77(1):59–91. doi: 10.1016/S0301-6226(01)00330-X. [DOI] [Google Scholar]
- Kampinga HH, Kanon B, Salomons FA, Kabakov AE, Patterson C. Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells. Mol Cell Biol. 2003;23:4948–4958. doi: 10.1128/MCB.23.14.4948-4958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamwanja LA, Chase CC, Gutierrez JA, Guerriero V, Olson TA, Hammond AC, Hansen PJ. Responses of bovine lymphocytes to heat-shock as modified by breed and antioxidant status. J Anim Sci. 1994;72(2):438–444. doi: 10.2527/1994.722438x. [DOI] [PubMed] [Google Scholar]
- Khalifa HH. Bioclimatology and adaptation of farm animals in a changing climate. Eaap Tech. 2003;7:15–29. [Google Scholar]
- Kishore A, Sodhi M, Khate K, Kapila N, Kumari P, Mukesh M. Selection of stable reference genes in heat stressed peripheral blood mononuclear cells of tropically adapted Indian cattle and buffaloes. Mol Cell Probe. 2013;27(3–4):140–144. doi: 10.1016/j.mcp.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Lacetera N, Bernabucci U, Ronchi B, Scalia D, Nardone A. Moderate summer heat stress does not modify immunological parameters of Holstein dairy cows. Int J Biometeorol. 2002;46(1):33–37. doi: 10.1007/s00484-001-0115-x. [DOI] [PubMed] [Google Scholar]
- Lacetera N, Bernabucci U, Scalia D, Ronchi B, Kuzminsky G, Nardone A. Lymphocyte functions in dairy cows in hot environment. Int J Biometeorol. 2005;50(2):105–110. doi: 10.1007/s00484-005-0273-3. [DOI] [PubMed] [Google Scholar]
- Lacetera N, Bernabucci U, Scalia D, Basirico L, Morera P, Nardone A. Heat stress elicits different responses in peripheral blood mononuclear cells from Brown Swiss and Holstein cows. J Dairy Sci. 2006;89(12):4606–4612. doi: 10.3168/jds.S0022-0302(06)72510-3. [DOI] [PubMed] [Google Scholar]
- Lacetera N, Bernabucci U, Basirico L, Morera P, Nardone A. Heat shock impairs DNA synthesis and down-regulates gene expression for leptin and Ob–Rb receptor in concanavalin A-stimulated bovine peripheral blood mononuclear cells. Vet Immunol Immunopathol. 2009;127(1–2):190–194. doi: 10.1016/j.vetimm.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Malayer JR, Hansen PJ. Differences between Brahman and Holstein cows in heat-shock induced alterations of protein synthesis and secretion by oviducts and uterine endometrium. J Anim Sci. 1990;68(1):266–280. doi: 10.2527/1990.681266x. [DOI] [PubMed] [Google Scholar]
- Marai IFM, Haeeb AAM. Buffalo's biological functions as affected by heat stress—a review. Livest Sci. 2010;127(2–3):89–109. doi: 10.1016/j.livsci.2009.08.001. [DOI] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Oksala NKJ, Laaksonen DE, Lappalainen J, Khanna S, Nakao C, Hänninen O, Sen CK, Atalay M. Heat shock protein 60 response to exercise in diabetes. Effects of α-lipoic acid supplementation. J Diabetes Complications. 2006;20:257–261. doi: 10.1016/j.jdiacomp.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Patir H, Upadhyay RC. Purification, characterization and expression kinetics of heat shock protein 70 from Bubalus bubalis. Res Vet Sci. 2010;88(2):258–262. doi: 10.1016/j.rvsc.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Paula-Lopes FF, Chase CC, Al-Katanani YM, Krininger CE, Rivera RM, Tekin S, Majewski AC, Ocon OM, Olson TA, Hansen PJ. Genetic divergence in cellular resistance to heat shock in cattle: differences between breeds developed in temperate versus hot climates in responses of preimplantation embryos, reproductive tract tissues and lymphocytes to increased culture temperatures. Reproduction. 2003;125(2):285–294. doi: 10.1530/rep.0.1250285. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signalling protein function and trafficking by the hsp90/hsp70 based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Silanikove N, Shapiro F, Shinder D. Acute heat stress brings down milk secretion in dairy cows by up-regulating the activity of the milk-borne negative feedback regulatory system. BMC Physiol. 2009;9:13. doi: 10.1186/1472-6793-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi M, Mukesh M, Kishore A, Mishra BP, Kataria RS, Joshi BK. Novel polymorphisms in UTR and coding region of inducible heat shock protein 70.1 gene in tropically adapted Indian zebu cattle (Bos indicus) and riverine buffalo (Bubalus bubalis) Gene. 2013;527:606–615. doi: 10.1016/j.gene.2013.05.078. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Gaffin SL, Pratt RE, Cullivam ML, Angel KG, Lilly CM. Effects of acute heat shock on gene expression by human peripheral blood mononuclear cells. J Appl Physiol. 2002;92:2208–2220. doi: 10.1152/japplphysiol.01002.2001. [DOI] [PubMed] [Google Scholar]
- Welch WJ. The mammalian stress response: cell physiology and biochemistry of stress proteins. In: Morimoto RI, Tissieres A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1990. pp. 223–278. [Google Scholar]
- Xiang-Hong J, Yan-Hong Y, Han-Jin X, Li-Long A, Ying-Mei X, Pei-Rong J, Ming L. Selection of reference genes for gene expression studies in PBMC from Bama miniature pig under heat stress. Vet Immunol Immunopathol. 2011;144(1–2):160–166. doi: 10.1016/j.vetimm.2011.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 39 kb)