Abstract
AIM: To evaluate the efficacy of a new hepatitis C virus (HCV) core antigen assay developed in China.
METHODS: After the determination of HCV infection, 49 serial samples were selected from 11 regular plasma donors in 5 different plasma stations. To compare the performance of HCV core antigen detection and HCV PCR, these samples were genotyped, and each specimen was analyzed by ELISA for the detection of HCV core antigen and by qualitative HCV PCR.
RESULTS: Among all of the sequential samples, the original 13 specimens were HCV RNA-negative, and 36 samples were HCV RNA-positive. Twenty-seven samples (75%) were HCV core antigen-positive from these HCV RNA-positive specimens. Conversely, 27 samples (93.1%) were found HCV RNA-positive in HCV core antigen-positive samples. Intervals between HCV RNA and HCV core antigen-positive, as well as between HCV core antigen-positive and HCV antibody-positive were 36.0 and 32.8 d, respectively.
CONCLUSION: This HCV core antigen assay, developed in China, is able to detect much of anti-HCV-negative, HCV RNA-positive preseroconversion window period (PWP) plasma donations.
Keywords: Hepatitis C virus, Core antigen, Anti-HCV, HCV RNA
INTRODUCTION
Before the introduction of detecting antibodies against recombinant and/or synthetic hepatitis C virus (HCV) antigens in blood centers, plasma stations, and hospitals post-transfusional non-A, non-B hepatitis, subsequently named after hepatitis C, was the major problem caused by HCV contamination of blood and blood products[1-3].
A sharp drop in post-transfusional hepatitis C was observed since the state-of-the-art screening assays of HCV antibodies were applied to blood donors[4,5]. However, patients of post-transfusion and sporadic hepatitis C infection were still reported for the presence of preseroconversion window period (PWP), in which antibody detection was not able to identify subjects in the early stage of infection. To date, specific antibodies against HCV have not been in spite of besides the existence of large numbers of HCV virus in the plasma of HCV-infected people. This phase may last up to 2 mo in immunocompetent subjects, and as long as 6 to 12 mo in immunodeficient patients[6]. The risk to a recipient of receiving a blood donation collected during the PWP for HCV has been estimated at 1/103 000 with a 95% confidence interval of 1/28 000 to 1/280 000 in USA from 1991 to 1993, 1/222 000 in France from 1993 to 1995, and 1/375 000 from 1996 to 1998[7,8].
To decrease the risk of HCV transmission from donors who are in the PWP at the time of their blood or plasma donation, nucleic acid amplification technology (NAT)-based tests have been applied to the detection of HCV RNA. Because NAT technology is a highly sensitive and specific technique, it is possible to identify viremic samples in which antibodies are not yet present and therefore reduce the window period to 15 to 20 d[9,10]. However, NAT tests require considerable skills, dedicated laboratory spaces, expensive reagents and long execution time. Taking into account the cost-efficacy ratio, the cost of NAT is too high to be used in most developing countries worldwide[11,12].
Recently, a new serologic test for the detection of the HCV core antigen in anti-HCV-negative serum or plasma has been developed and is commercially available in China. This ELISA assay can be used as a screening assay for blood or plasma donations during PWP as an alternative to NAT. The aim of the present study was to compare the efficacy of this assay with a qualitative HCV RNA detection, and estimate the reduction in the PWP from that observed with HCV antibody assay by using a set of serial plasma samples collected in Hunan Province, Southern China.
MATERIALS AND METHODS
Plasma samples
Plasma samples were collected from 5 plasma stations located in different counties of Hunan Province, Southern China (Huaihua, Xinhua, Liling, Junshan, and Quyuan). These 5 plasma stations have been operating plasma collection for more than 5 years. During this period, 48 000 plasma donors have been registered, and among them 27 500 plasma donors have been continuously donating plasma for more than 2 years and donated at least 15 times during this period. Two milliliters plasma aliquots for each donation were preserved in a freezer in the plasma station. Before collection, each plasma donor was tested with two different third-generation HCV Ab ELISA assays. The first one is domestic third-generation HCV Ab ELISA assay (Wantai Biological Pharmacy, Beijing, PR China), and positive samples [sample-to-cut-off ratio (S/CO) is higher than 1] were also tested by the other imported third-generation ELISA assay (Abbott, USA). Double-positive plasma samples were selected and determined as HCV Ab-positive, that means HCV Ab is present in the plasma by that time. According to the availability of the specimens that were kept frozen since their collection, a total of 49 serial plasma specimens (3-6 per patient) from 11 plasma donors were selected for the determination of HCV Ab-positive specimen.
Qualitative detection of HCV RNA
For qualitative detection of HCV RNA, viral RNA was extracted from 50 μL of serum or plasma by using the QiaAmp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Subsequently, the nested cDNA RT-PCR assay was used for detecting the presence of HCV RNA (Sino-American Biotechnology, Luoyang, Henan Province, China) following the manufacturer’s instructions. The conservative sequence in the end of the 5’-noncoding region (NCR) of the HCV genome was amplified and analyzed using the two pairs of nested primers supplied with the kit. The sensitivity and specificity of the qualitative detection of HCV RNA detection (Sino-American Biotechnology) were 99% and 95%, respectively.
Determination of HCV genotype
The HCV genotype was determined by the method reported by Du et al[13]. A 257-bp fragment from 5’ noncoding region (5’-NCR) of cDNA was amplified by RT-PCR. The product was digested using a combination of three restriction endonuclease BHH’ (BsrBI, Hea IIand HinfI), using restriction endonuclease BstU I, and using restriction endonuclease Hae III, respectively. The test results were interpreted according to the algorithm described by the authors[13].
HCV core antigen assay
HCV core antigen was detected with an HCV Ag ELISA assay (Kinda Gene, Changsha, Hunan Province, China) by strictly following the manufacturer’s instructions. This assay is based on a two-step sandwich principle. Briefly, murine monoclonal antibodies recognizing different epitopes of HCV core antigen were coated on microwells of the solid phase and were conjugated with horseradish peroxidase. One hundred microliters of serum or plasma specimen was added to the microwells combined with the same value of specimen diluent, the microplate was incubated at 37°C in waterbath with vibration for 90 min. After washing 5 times, 200 μL of conjugate (monoclonal anti-HCV core protein conjugated with HRP) was added and the microplate was incubated at 37°C in waterbath for 30 min. After washing as described above, 100 μL of substrate solution A (350 mmol/L sodium acetate, 10 mmol/L sodium citrate, 300 mL/L H2O2) and 100 μL of substrate solution B (1.3 mmol/L TMB, 1 mmol/L EDTA-Na2, 10 mmol/L citric acid, 105 mL/L glycerol) were added into microwells, the microplate was placed at 37°C in dark for 10-15 min, then 50 μL of stop solution (1 mol/L H2SO4) was added. The absorbance (A450) of the microwells was determined at ultraviolet wavelength 450 nm within 10 min. The test was considered valid if the A450 of positive control was greater than 0.6 and negative control was less than 0.1. The results were expressed by using the S/CO ratios: the A450 of specimen was divided by the cutoff value. The cutoff value was determined based on the distribution of absorbance values of the specimens from healthy subjects. In present study, the cutoff value was tentatively calculated by adding 0.05 (3 ×SD) to the mean A450 of healthy subjects’ sera (0.068, n = 400), i.e. the cutoff value was 0.118. S/CO ratios ≥ 1.0 were interpreted as positive.
Statistical analysis
The correlations between HCV core antigen ELISA assay and qualitative HCV RNA assay were analyzed by McNemar χ² test and kappa test. The statistical package SAS8.0 (SAS Institute, Cary, NC) was used for data analysis. Two-sided P ≤ 0.05 was considered statistically significant.
RESULTS
Individual plasmas and HCV genotypes
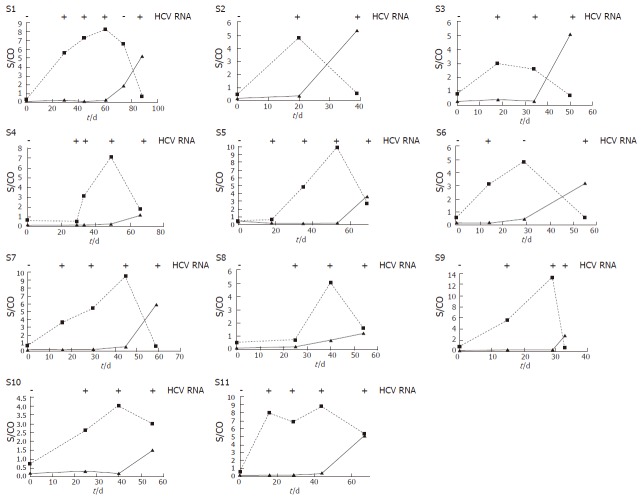
Forty-nine sequential samples from 11 anti-HCV-positive plasma donors (S1-S11) were selected and kept in frozen without thawing. Of them, the earliest 11 specimens were negative for both HCV RNA and HCV core antigen and defined as d 0 for plasma donation. For the other 38 plasma samples, 29 and 36 specimens are positive for HCV core antigen and HCV RNA, respectively. Twenty-seven of 36 (75%) HCV RNA-positive specimens were HCV core antigen-positive. Twenty-seven (93.1%) of 29 HCV core antigen-positive samples were found to be HCV RNA-positive. The average time for HCV RNA to be became detectable was 20.4 d (range, 14-29 d), for HCV core antigen was 23.7 d (range, 14-40 d) and for HCV Ab was 56.5 d (range, 33-74 d). Genotyping results showed that HCV genotype of these plasma samples mainly was 1b (9/11) or 1a (2/11) and basically covered the major prevalent genotype in China (Table 1).
Table 1.
Days of the first detection for HCV RNA, HCV core antigen and HCV Ab and the intervals between them
| Case number | Genotype | HCV RNA | HCV core antigen | HCV Ab | HCV core antigen to HCV Abs | HCV RNA to HCV Abs | HCV RNA to HCV core antigen |
| S1 | 1b | 29 | 29 | 74 | 45 | 45 | 0 |
| S2 | 2a | 20 | 20 | 39 | 19 | 19 | 0 |
| S3 | 1b | 18 | 18 | 50 | 32 | 32 | 0 |
| S4 | 1b | 29 | 33 | 66 | 33 | 37 | 4 |
| S5 | 1b | 18 | 35 | 69 | 34 | 51 | 17 |
| S6 | 1b | 14 | 14 | 55 | 41 | 41 | 0 |
| S7 | 1b | 16 | 16 | 59 | 43 | 43 | 0 |
| S8 | 1b | 25 | 40 | 54 | 14 | 29 | 15 |
| S9 | 1b | 15 | 15 | 33 | 18 | 18 | 0 |
| S10 | 1b/2a | 25 | 25 | 55 | 30 | 30 | 0 |
| S11 | 1b | 16 | 16 | 67 | 51 | 51 | 0 |
| Average days | 20.4 | 23.7 | 56.5 | 32.7 | 36 | 3.3 |
Qualitative analysis of HCV RNA and HCV core antigen
According to our results, the mean times from the first detection of HCV RNA and HCV core antigen to HCV Abs were evaluated in 11 plasma donors (Table 1). Times from the first HCV RNA and HCV core antigen-positive to HCV Ab-positive ranged from 19 to 51 (mean, 36.0) d and 14 to 51 (mean 32.7) d, respectively. The times from HCV RNA-positive to HCV core antigen-positive were 4, 17, and 15 d in donors S4, S5, and S8, respectively. HCV RNA and HCV core antigen were both detected on the same specimens from the other 8 donors, so the average interval was 3.3 d. Table 2 shows the comparative results of qualitative HCV RNA and HCV core antigen detection. Statistic analysis showed no significant difference (P = 0.07) between HCV RNA detection and HCV core antigen assay, and a good coherence was found (K = 0.51) between them. The evolution of three HCV markers-HCV RNA, HCV core antigen, and HCV Ab-in 11 plasma donors is illustrated in Figure 1.
Table 2.
Comparative results of HCV RNA and HCV core antigen detection in 49 serial samples from 11 plasma donors
|
HCV RNA |
|||
| HCV core antigen | Negative | Positive | Total |
| Negative | 11 | 9 | 20 |
| Positive | 2 | 27 | 29 |
| Total | 13 | 36 | 49 |
Figure 1.
Evaluations of HCV core antigen ( ■), and HCV antibody ( ▲) in eleven donations (S1-S11) within preseroconversion window period. HCV core antigen assay and HCV antibody S/CO values are plotted versus the time of sample acquisition. (First samples = d 0).
DISCUSSION
HCV belongs to the Flaviviridae family, and its genome is packaged into an icosahedral capsid (or core)[14]. The capsid is composed of the HCV core protein, a structural viral protein encoded by the 5’ end of the HCV open reading frame. The HCV core protein is highly conserved and antigenic. So, HCV core protein can induce strong specific cellular and humoral responses, and probably plays a pivotal role in the pathogenesis of HCV infection[15,16].
An ELISA assay to detect HCV core antigen in peripheral blood of patients with HCV has been developed after the anti-HCV core antigen monoclonal antibody is available[17,18]. Qualitative and quantitative ELISA kit (Ortho-Clinical Diagnostics, Johnson & Johnson Company) have been successively developed for assessment of HCV core antigen and commercially available out of China, such as in Thailand and Italy, to detect HCV core antigen[19,20]. A number of reports have suggested that these assays could be used to reduce the serological preseroconversion window period, measure HCV replication, and monitor the response to antiviral therapy[21-23].
In this study, a new commercially available ELISA assay for the detection of “free” HCV core antigen developed by Kinda Gene Company (Changsha, Hunan Province, PR China) was evaluated. Good correlation (75%) was observed between assays of HCV core antigen and qualitative HCV RNA, in accordance with 71% in the early infection phase[22], but the positive rate of the HCV core antigen ELISA was lower than previously reported rates of 87%, 88%, or 94% for other PWP specimens[24,25]. The discrepancy is likely due to the selection of early PWP plasma samples with lower HCV titers, or to the use of a highly sensitive qualitative RT-PCR method for single donations under well-controlled conditions[8,24,25].
Detection of HCV antibodies is commonly used in blood centers and plasma stations to reduce the transmission of HCV in China. To reduce the risk of PWP donation, the European Medicinal Evaluation Agency (EMEA) has introduced NAT-based test for detection of plasma pools in the manufacture of blood products for HCV RNA since 1999[22]. The highest sensitivity of NAT technique may also enable it to be applied to pools of sera. Except of the advantage of the highest sensitivity, current limits of NAT technique which are the lack of automation, a high cost-efficacy ratio, and the risk of false-positive result have led it not to be broadly used in most of under-developed region, such as Africa, Asia, and Latin America. On the other hand, serological detection of HCV core antigen may be an alternative to NAT-based methods. The NAT-based method involve expensive reagents and long execution times, aspects that have a significant effect on the final cost of each test, which is around US$10 per test when the test is performed in diagnostic laboratories. Therefore, a quick, inexpensive, sensitive, and specific test is clearly needed to identify potentially infective blood units that have not been identified by specific antibody tests. The new serologic test for the detection of the HCV core antigen in anti-HCV-negative serum or plasma developed and commercially available in China appears to decrease the cost of the test, which is around one-tenth of the NAT-based method. So, the HCV core antigen ELISA can be easily performed in clinical laboratories, it is simple, reliable, rapid and non-expensive. Our data showed that the total window period estimated at 56.5 d could be reduced to 23.7 d by testing for HCV core antigen. Hence, HCV core antigen ELISA is able to detect most anti-HCV-negative, HCV RNA-positive PWP donations. Because of the limitation of sample numbers, no difference of the variables studied, such as S/CO ratio, size of PWP, was found between HCV two genotypes prevailed in China.
In conclusion, our study shows that the HCV core antigen assay is cheap, easily performed, and compatible to equipments now used in blood centers and plasma stations; its application would lead to a significant improvement in transfusion safety and would save more money and time in comparison to the current NAT. Moreover, the development of the newer, more-sensitive, quantitative second version of the assay will greatly improve the performance of HCV core antigen assay which may be the good alternative of NAT-based assay in most countries of the world, including China.
Footnotes
Supported by the National Key Technologies R&D Program of China during the 10th Five-Year Plan, No. 2001BA705B06; and National High Technology Research and Development Program of China (863 Program), No. 2006AA020907
S- Editor Liu Y L- Editor Kumar M E- Editor Che YB
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Couroucé AM. Development of screening and confirmation tests for antibodies to hepatitis C virus. Curr Stud Hematol Blood Transfus. 1998;(62):64–75. [PubMed] [Google Scholar]
- 3.van der Poel CL. Hepatitis C virus and blood transfusion: past and present risks. J Hepatol. 1999;31 Suppl 1:101–106. doi: 10.1016/s0168-8278(99)80384-5. [DOI] [PubMed] [Google Scholar]
- 4.Donahue JG, Muñoz A, Ness PM, Brown DE, Yawn DH, McAllister HA, Reitz BA, Nelson KE. The declining risk of post-transfusion hepatitis C virus infection. N Engl J Med. 1992;327:369–373. doi: 10.1056/NEJM199208063270601. [DOI] [PubMed] [Google Scholar]
- 5.Wang YJ, Lee SD, Hwang SJ, Chan CY, Chow MP, Lai ST, Lo KJ. Incidence of post-transfusion hepatitis before and after screening for hepatitis C virus antibody. Vox Sang. 1994;67:187–190. doi: 10.1111/j.1423-0410.1994.tb01657.x. [DOI] [PubMed] [Google Scholar]
- 6.van der Poel CL, Cuypers HT, Reesink HW. Hepatitis C virus six years on. Lancet. 1994;344:1475–1479. doi: 10.1016/s0140-6736(94)90293-3. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber GB, Busch MP, Kleinman SH, Korelitz JJ. The risk of transfusion-transmitted viral infections. The Retrovirus Epidemiology Donor Study. N Engl J Med. 1996;334:1685–1690. doi: 10.1056/NEJM199606273342601. [DOI] [PubMed] [Google Scholar]
- 8.Couroucé AM, Le Marrec N, Bouchardeau F, Razer A, Maniez M, Laperche S, Simon N. Efficacy of HCV core antigen detection during the preseroconversion period. Transfusion. 2000;40:1198–1202. doi: 10.1046/j.1537-2995.2000.40101198.x. [DOI] [PubMed] [Google Scholar]
- 9.Müller-Breitkreutz K, Baylis SA, Allain J. Nucleic acid amplification tests for the detection of blood-borne viruses. 5th EPFA/NIBSC Workshop, Amsterdam 1998. Vox Sang. 1999;76:194–200. doi: 10.1159/000031050. [DOI] [PubMed] [Google Scholar]
- 10.Seme K, Poljak M. Evaluation of the Amplicor HCV test: experiences after 1 year of routine use in a diagnostic laboratory. Infection. 1996;24:140–143. doi: 10.1007/BF01713321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poljak M, Seme K, Koren S. Evaluation of the automated COBAS AMPLICOR hepatitis C virus PCR system. J Clin Microbiol. 1997;35:2983–2984. doi: 10.1128/jcm.35.11.2983-2984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth WK, Weber M, Seifried E. Feasibility and efficacy of routine PCR screening of blood donations for hepatitis C virus, hepatitis B virus, and HIV-1 in a blood-bank setting. Lancet. 1999;353:359–363. doi: 10.1016/S0140-6736(98)06318-1. [DOI] [PubMed] [Google Scholar]
- 13.Du SC, Sun NX, Fan XF, Qiu GH, You P, Zhang YX, Wei L. Hepatitis C virus (HCV) 5' NCR genotype research and 3b sequencing analysis in China. Zhonghua Jianyan Yixue Zazhi. 2004;27:683–686. [Google Scholar]
- 14.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 15.Lai MM, Ware CF. Hepatitis C virus core protein: possible roles in viral pathogenesis. Curr Top Microbiol Immunol. 2000;242:117–134. doi: 10.1007/978-3-642-59605-6_6. [DOI] [PubMed] [Google Scholar]
- 16.Nelson DR, Marousis CG, Davis GL, Rice CM, Wong J, Houghton M, Lau JY. The role of hepatitis C virus-specific cytotoxic T lymphocytes in chronic hepatitis C. J Immunol. 1997;158:1473–1481. [PubMed] [Google Scholar]
- 17.Aoyagi K, Ohue C, Iida K, Kimura T, Tanaka E, Kiyosawa K, Yagi S. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999;37:1802–1808. doi: 10.1128/jcm.37.6.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orito E, Mizokami M, Tanaka T, Lau JY, Suzuki K, Yamauchi M, Ohta Y, Hasegawa A, Tanaka S, Kohara M. Quantification of serum hepatitis C virus core protein level in patients chronically infected with different hepatitis C virus genotypes. Gut. 1996;39:876–880. doi: 10.1136/gut.39.6.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Netski DM, Wang XH, Mehta SH, Nelson K, Celentano D, Thongsawat S, Maneekarn N, Suriyanon V, Jittiwutikorn J, Thomas DL, et al. Hepatitis C virus (HCV) core antigen assay to detect ongoing HCV infection in thai injection drug users. J Clin Microbiol. 2004;42:1631–1636. doi: 10.1128/JCM.42.4.1631-1636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabrizi F, Lunghi G, Aucella F, Mangano S, Barbisoni F, Bisegna S, Vigilante D, Limido A, Martin P. Novel assay using total hepatitis C virus (HCV) core antigen quantification for diagnosis of HCV infection in dialysis patients. J Clin Microbiol. 2005;43:414–420. doi: 10.1128/JCM.43.1.414-420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valcavi P, Medici MC, Casula F, Arcangeletti MC, De Conto F, Pinardi F, Calderaro A, Chezzi C, Dettori G. Evaluation of a total hepatitis C virus (HCV) core antigen assay for the detection of antigenaemia in anti-HCV positive individuals. J Med Virol. 2004;73:397–403. doi: 10.1002/jmv.20105. [DOI] [PubMed] [Google Scholar]
- 22.Agha S, Tanaka Y, Saudy N, Kurbanov F, Abo-Zeid M, El-Malky M, Khalaf M, Ohta N, Yoshizawa H, Mizokami M. Reliability of hepatitis C virus core antigen assay for detection of viremia in HCV genotypes 1, 2, 3, and 4 infected blood donors: a collaborative study between Japan, Egypt, and Uzbekistan. J Med Virol. 2004;73:216–222. doi: 10.1002/jmv.20078. [DOI] [PubMed] [Google Scholar]
- 23.Icardi G, Bruzzone B, Gota F, Torre F, Giannini E, Massone L, Li Bassi A, Lai PL, Picciotto A, Ansaldi F. A new assay for hepatitis C virus (HCV) core antigen detection: an alternative to nucleic acid technologies in positive or indeterminate anti-HCV subjects. Ann Ig. 2003;15:863–870. [PubMed] [Google Scholar]
- 24.Lee SR, Peterson J, Niven P, Bahl C, Page E, DeLeys R, Giordano-Schmidt D, Baggett D, Green G. Efficacy of a hepatitis C virus core antigen enzyme-linked immunosorbent assay for the identification of 'window-phase' blood donations. Vox Sang. 2001;80:19–23. doi: 10.1046/j.1423-0410.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- 25.Peterson J, Green G, Iida K, Caldwell B, Kerrison P, Bernich S, Aoyagi K, Lee SR. Detection of hepatitis C core antigen in the antibody negative 'window' phase of hepatitis C infection. Vox Sang. 2000;78:80–85. doi: 10.1159/000031155. [DOI] [PubMed] [Google Scholar]