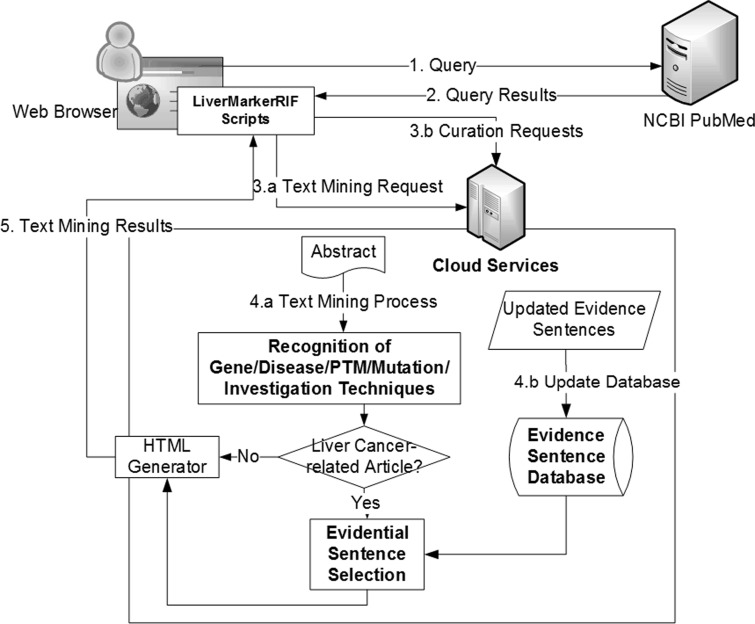
Figure 3.
The system workflow of LiverCancerMarkerRIF. After installing LiverCancerMarkerRIF, it monitors the query sent to PubMed (step 1 and 2) and analyzes the returned results to extract the target text, such as the title and abstract. In step 3.a, the extracted text is sent to the cloud server to recognize biomedical entities. In step 4.a, in addition to recognizing entities, the curated evidential sentences for the article are further extracted from our database and combined with the entity recognition results if the text contained liver cancer-related terms. The results are finally sent back to the LiverCancerMarkerRIF in the client side to augment the original content. If some evidential sentences are curated by the curator, LiverCancerMarkerRIF will issue a curation request (step 3.b) and update our server-side database (step 4.b).