Fig. 1.
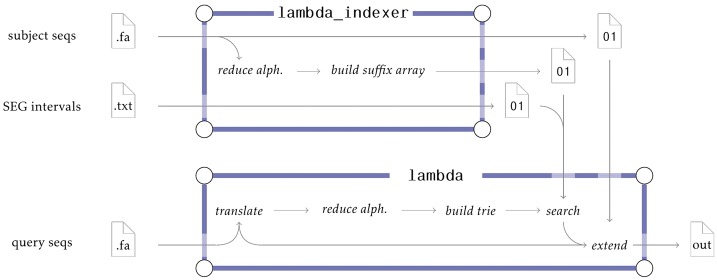
Pipeline of Lambda (BlastX-mode). The lambda_indexer takes as input a file with the subject sequences and optionally an interval file computed by seqmasker to create the index for seed identification. The query sequences are translated from DNA to amino acid alphabet, reduced and then converted into a search trie. Afterwards, the search trie is used together with the pre-computed index to find candidate regions, which are then extended and verified
