Abstract
Elafin (peptidase inhibitor 3 [PI3]) and its biologically active precursor, pre-elafin, are neutrophil serine proteinase inhibitors with an important role in preventing excessive tissue injury during inflammatory events. Recently, we reported an association between single-nucleotide polymorphism (SNP) rs2664581 in the PI3 gene, increased risk of acute respiratory distress syndrome (ARDS) and pre-elafin circulating levels. This study aims to validate the legitimacy of this association by using a cohort of patients who met the criteria for systemic inflammatory response syndrome and were at risk of developing ARDS (n = 840). A comprehensive functional study of SNPs in PI3 gene was also performed. Luciferase assays and electrophoretic mobility shift assays were conducted to determine the functional relevance of promoter region variants. The effect of the coding SNP rs2664581 on the neutrophil elastase inhibitory activity and transglutaminase binding properties of pre-elafin was also investigated. The variant allele of rs2664581 (C) was significantly associated with increased ARDS risk, mainly among subjects with sepsis (odds ratio = 1.44; 95% confidence interval = 1.04–1.99; P = 0.0276, adjusted by age, sex, and Acute Physiology and Chronic Health Evaluation III). Pre-elafin recombinant protein carrying the amino acid change associated with rs2664581 (Thr34Pro, mutant protein [MT]) had greater capacity to undergo transglutaminase-mediated cross-linking to immobilized fibronectin than wild-type protein in vitro (P < 0.003). No differences were observed in the neutrophil elastase inhibitory activities of wild-type versus MT proteins. In addition, the risk allele–promoter construct had significantly lower cytokine-induced transcriptional activity. Electrophoretic mobility shift assay results indicated a differential binding of nuclear proteins to the G and A alleles of SNP −338G > A. Our results confirm the association between SNP rs2664581 and enhanced risk of ARDS, further supporting the role of PI3 in ARDS development. SNPs in the PI3 locus may act synergistically by regulating PI3 gene expression and pre-elafin biological functions.
Keywords: acute respiratory distress syndrome, elafin, single-nucleotide polymorphism, genetic association, replication
Clinical Relevance
Our present study confirms previous association between peptidase inhibitor 3 (PI3) polymorphisms and acute respiratory distress syndrome (ARDS) risk supporting earlier evidence of the role of pre-elafin in the pathophysiology of ARDS. Our results also show that genetic variation in PI3 gene differentially regulates its expression, and may have important consequences in protein function. Altogether, these modifications might have a significant impact on susceptibility to ARDS development.
Human neutrophil elastase (HNE) has emerged as a key effector in lung injury (1). This serine proteinase is secreted by neutrophils, and participates as a part of the host defense against microbial pathogens by facilitating neutrophil migration through the tissues to the inflammatory site and killing bacteria and fungi (2, 3). However, when the inflammatory process is extreme, excessive NE activity has deleterious consequences for host tissues (4). For example, uncontrolled NE activity in the lung leads to increased alveolar–capillary permeability and an influx of protein-rich fluid into the alveolar space, which initiates and propagates pulmonary injury (1, 4–6). There is now good evidence that NE contributes significantly to the pathogenesis of chronic obstructive pulmonary disease, cystic fibrosis, acute respiratory distress syndrome (ARDS), and pulmonary fibrosis (7). Thus, novel therapeutics that adequately control NE activity have great potential to protect the lung from injury during intense inflammatory responses. Pre-elafin (elafin, skin-derived antileukoprotease, trappin-2, peptidase inhibitor 3 [PI3]) is a canonical inhibitor belonging to the chelonianin family of proteinase inhibitors (8). PI3 encodes a secreted 9.9-kD protein consisting of 95 amino acids (9, 10). Pre-elafin is classified as an “alarm” antiproteinase, because it is synthesized and secreted by cells at sites of inflammation in the lung in response to the same primary stimuli (IL-1β, TNF-α, and also NE) that drive the initial inflammatory response (11–13), providing a local and efficient defense against deleterious serine proteinase activity. Pre-elafin has been proved to have significant protective effects against lung inflammation and lung injury induced by HNE, LPS, or Pseudomonas aeruginosa in several murine acute lung injury (ALI) models (14–16). In addition to its role as a proteinase inhibitor, many other biological activities have been attributed to this molecule, including antibacterial, antifungal, antiviral, antiinflammatory, and immunomodulatory functions (17–19). Thus, pre-elafin contributes in important ways to host defense and tissue homeostasis, which augments considerably the therapeutic potential of this protein in inflammatory respiratory diseases. Our previous genome-wide expression analysis showed that PI3 gene expression is down-regulated during the acute stage of ARDS, and this correlates well with plasma pre-elafin levels (20). Recently, we found that single-nucleotide polymorphism (SNP) rs2664581, located in exon 2 of PI3 gene, is associated with increased ARDS risk and pre-elafin circulating levels (21). This SNP results in an amino acid substitution (Thr34Pro) within the N-terminal (cementoin) domain of pre-elafin, which anchors the molecule to extracellular matrix (ECM) proteins (22–25). This change may affect the ability of pre-elafin to be cross-linked by transglutaminase (TGase) to ECM proteins, which increases pre-elafin’s capacity to protect ECM proteins from proteolytic degradation by neutrophil serine proteinases. Because of the high linkage disequilibrium (LD) between SNP rs2664581 associated with ARDS development and the remaining SNPs in the PI3 locus (21), true causative variations remain to be identified, together with their specific effects on PI3 expression, and/or pre-elafin biological functions. In the present study, we attempted to validate previous associations between genetic variants in PI3 gene (SNP rs2664581) and ARDS development. To gain insights into the mechanism leading to PI3 ARDS–predisposing effects, we performed a thorough functional characterization of all SNPs in the PI3 promoter and the coding SNP rs2664581. We hypothesized that PI3 polymorphisms may act synergistically, with multiple effects at both the transcriptional and post-translational levels on pre-elafin expression and function, to contribute significantly to the development of ARDS.
Material and Methods
Study Populations
We studied 840 patients included in the Harborview Medical Center (HMC) systemic inflammatory response syndrome (SIRS) cohort, and admitted to the intensive care units (ICUs) of HMC (Seattle, WA) from December 2006 to December 2010. Details of patient population have been described previously (26, 27). Demographic data, severity of illness scores, and clinical information were recorded on ICU admission. Enrolled patients who fulfilled the Berlin criteria for mild, moderate, or severe ARDS (28) (corresponding to previous consensus criteria for ALI, with a positive end-expiratory pressure [PEEP] ≥ 5 cm H2O) (29) were considered as cases. At-risk patients who did not meet criteria for the development of ARDS during the ICU hospitalization were classified as control subjects. To reduce the potential confounding effect of ethnic background, only white patients were analyzed. The Institutional Review Board and/or Human Subjects Committee reviewed and approved the study. The study design and patient selection flow diagram is illustrated in Figure E1 in the online supplement.
Genotyping and Quality Control
Genomic DNA was isolated using the Qiagen QIAamp DNA Blood Midi Kit (Qiagen, Valencia, CA). Patients were genotyped by the 5′ nuclease assay (TaqMan; Life Technologies, Carlsbad, CA) using the Biomark platform (Fluidigm, San Francisco, CA). Genotyping was performed by laboratory personnel without the knowledge of case–control status. The overall genotyping success rate was above 99%. For quality control, samples from nonwhite subjects as well as from subjects with missing genotyping or clinical data were excluded from analysis.
Identification of Polymorphisms in PI3 Promoter and LD Block Analysis
Polymorphisms in PI3 promoter gene were identified by sequencing 2 kb of the 5′ untranslated regions using DNA samples from 29 nonrelated, healthy white subjects, as previously described (21). Resequencing data were used to determine the LD structure of the PI3 promoter as described previously (30, 31).
Design and Construction of PI3 Promoter Haplotype-Reporter Plasmids
To obtain the constructs for the luciferase assay, the PI3 gene promoter (from −2,378 to +1) was amplified by PCR using genomic DNA from subjects carrying each haplotype. The primers used were: sense, 5′-atatatGTCGACctgggtgacaagagcgaaattcc-3′; antisense, 5′-ggagcaGGGCCCttcttaatgttcttagcatcggccatggtgtcaggaaggtgtttg-3′, (note capitals representing restriction sites for SalI and ApaI at the 3′ of each primer, respectively). The resulting PCR products were subsequently digested with SalI and ApaI and cloned in the luciferase reporter vector, pEZX-MT01 (GeneCopoeia, Inc., Germantown, MD) yielding the plasmids pPI3-h1 and pPI3-h2 (Figure E2), where the expression of firefly luciferase hluc gene is under the control of wild-type (WT) and mutant (MT) PI3 promoters, respectively. All reporter constructs were sequenced to confirm the fidelity of the amplification.
Tissue Culture, Transient Transfection, and Luciferase Reporter Assay
A human alveolar basal epithelial cell line (A549) was obtained from American Tissue Culture Collection (Manassas, VA) and maintained as monolayers in F-12K medium (American Tissue Culture Collection) supplemented with 100 U/ml penicillin and 100 U/ml streptomycin and 10% (vol/vol) FBS at 37°C in a 5% CO2 atmosphere. Cells were transfected at approximately 90–95% confluence with 1 μg of each reporter plasmid using Lipofectamine 2,000 transfection reagent in Opti-MEM serum-free medium (Life Technologies) according to the manufacturer’s instruction. The renilla luciferase hRluc gene in plasmids pPI3-h1 and pPI3-h2 was used as an internal transfection control. At 24 hours after transfection, cells were stimulated with IL-1β and TNF-α (20 ng/ml) for 24 hours. Cytoplasmic extracts were then prepared and luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to manufacturer’s protocol and quantified using the SpectraMax M5 Microplate Spectrophotometer (Molecular Devices, Sunnyvale, CA). Results were normalized for internal renilla luciferase activity and expressed as relative luciferase activity. Three independent experiments were performed in triplicate wells per assay.
Electrophoretic Mobility Shift Assay
Nuclear extracts were obtained from A549 cells incubated in the presence or absence of 20 ng/ml of IL-1β for 24 hours using the NE-PER nuclear extraction reagent (Thermo Scientific, Rockford, IL). Nuclear proteins were quantified using the Micro BCA Protein Assay kit (Thermo Scientific, IL). Biotin end-labeled synthetic oligonucleotides were purchased from Life Technologies. Complimentary oligos were mixed at equimolar concentrations, incubated at 95°C for 5 minutes, and then annealed by slow cooling to room temperature for 45–60 minutes to form double-stranded probe. Three different biotin-labeled oligos (and the corresponding unlabeled cold competitors) were used in our experiments: G oligo, 5′-biotin-TAACCTTCGGTGATTCCTTTCT-3′ containing the major allele of SNP −338 (G); A oligo, 5′-biotin-TAACCTTCGATGATTCCTTTCT-3′ containing the minor allele of SNP −338 (A); and NF-κB oligo, 5′-biotin-AGTTGAGGGGACTTTCCCAGGC-3′ containing the NF-κB consensus sequence. Binding reactions were performed using the LightShiftTM Chemiluminescent Electrophoretic Mobility Shift Assay (EMSA) kit (Thermo Scientific). Briefly, nuclear protein extracts (2 μg) were incubated in buffer containing 10 mM Tris, 1 mM dithiothreitol (pH 7.5), 50 mM KCl, (pH 7.5), 50 ng/μl of poly (dI-dC), and 16 fmol of the biotinylated oligonucleotides for 30 minutes at room temperature. For competition experiments, unlabeled probes (8 pmol) were added to the reaction in the presence of 200 mM KCl 20 minutes before the addition of the labeled probes to confirm protein-binding specificity. The DNA:protein complexes were resolved on a 4.5% nondenaturing polyacrylamide gel in 1× TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) at 85 V for 2 hours at 4°C and then transferred to a Hybond-N+ nylon membrane (GE Healthcare Lifescience, Buckinghamshire, UK). The transferred complexes were cross-linked for 15 minutes on a ultraviolet transilluminator equipped with 312-nm bulbs. The nuclear proteins:biotin-labeled DNA complexes were visualized using a horseradish peroxidase–conjugated streptavidin antibody and a chemiluminiscent substrate (Chemiluminiscent Nucleic Acid Detection Module; Thermo Scientific) according to manufacturer’s instruction. Bands were detected using a ChemiDoc XRS+ System with Image Lab Software (Bio-Rad Laboratories Inc., Hercules, CA).
Chemicals and Enzymes
To investigate the effect of SNP rs2664581 on pre-elafin biological functions, we designed a pre-elafin protein variant (MT), in which the amino acid, Proline, at position 34 is replaced with Threonine mimicking the amino acid change associated with SNP rs2664581. Recombinant WT and MT pre-elafin were obtained from Abnova Taiwan Corp. (Tapei City, Taiwan). HNE and pig liver TGase were purchased from Athens Research and Technology (Athens, GA) and Sigma-Aldrich (St. Louis, MO), respectively. The quenched fluorogenic substrate that is specific for HNE (MeoS-Ala-Ala-Pro-Val-AFC) was purchased from Enzyme Systems Products (Livermore, CA). Color Reagent A (stabilized peroxide solution) and B (stabilized chromogen solution) were from R&D Systems (Minneapolis, MI). Rabbit anti-human elafin antibody (FL-117) and goat anti-rabbit IgG conjugated to horseradish peroxidase were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
NE Assay
The inhibitory activities of recombinant pre-elafin proteins were compared by incubating equimolar concentrations (0.05, 0.1, 0.5, 1, 5, 10, 20, 30, 50, and 100 nM) of these inhibitors with HNE (50 nM) for 60 minutes at 37°C and then measuring residual HNE activity using a quenched fluorogenic substrate specific for HNE (MeoS-Ala-Ala-Pro-Val-AFC), as previously described (32). Dose–response curves were constructed and the half-maximal inhibitory concentration of each inhibitor was determined.
Quantification of WT and MT Pre-Elafin Recombinant Proteins Covalently Bound to Immobilized Fibronectin by TGase Using an ELISA
The binding of WT and MT pre-elafin proteins to fibronectin was assessed by ELISA as previously described (33). Briefly, 96-well Costar ELISA microplates (Corning, Inc., Corning, NY) were coated with purified plasma human fibronectin (1.5 μg/well in 100 μL PBS) overnight at 4°C. After blocking free binding sites with PBS containing 1% BSA, recombinant pre-elafin proteins (0.5, 1, and 1.5 × 10−7 M) were added to triplicate wells together with TGase (1.25 × 10−7 M) and incubated at 37°C for 1 hour. Specificity of covalent binding of pre-elafin proteins to fibronectin was measured in additional triplicate wells by omitting TGase for all concentrations of inhibitor tested. Covalently linked pre-elafin proteins were detected with rabbit anti–pre-elafin polyclonal antibody, followed by peroxidase-conjugated goat anti-rabbit antibody. Peroxidase activity was measured by adding a chromogenic substrate (Color Reagents A and B from R&D Systems) at 450 nm using a SpectraMax Microplate Spectrophotometer.
Bioinformatics
The prediction of transcription factor (TF) binding sites (TFBSs) in the PI3 promoter was performed using Genomatix MatInspector 8.0 (34) that identifies TFBSs in nucleotide sequences using a large library of weight matrices and Transcription Element Search System (http://www.cbil.upenn.edu/cgi-bin/tess/tess), which can identify binding sites using site or consensus strings and positional weight matrices from the Transcription Factor Database, JASPAR, Information Matrix Database, and Computational Biology and Informatics Laboratory-GibbsMat databases. Searches for TFBSs were made with the default parameters for both software programs. Jalview (http://www.jalview.org/) and AGADIR software (http://agadir.crg.es/) were used to perform alignment and to visualize the effect of SNP rs2664581 on the secondary structure of the cementoin domain of the protein.
Statistical Analyses
All statistical analyses were performed using the SAS statistical software package (version 9.1; SAS, Cary, NC). A P value less than 0.05 was considered to be statistically significant. The demographic variables between patients with ARDS and control subjects were tested by Fisher’s exact test for categorical variables and by Student’s t test for continuous variables. We used SAS/Genetics PROC ALLELE test to calculate the allele frequencies, to test the deviation from Hardy-Weinberg equilibrium. We used multivariate logistic regression to estimate the genotype-specific odds ratio (OR) and 95% confidence interval (CI) for ARDS susceptibility. Covariates were chosen based on the potential risk for ARDS development, including age, sex, and Acute Physiology and Chronic Health Evaluation (APACHE) III score. Logistic regression was also used in the comparison of the binding of WT and MT pre-elafin proteins to fibronectin. One-way ANOVA was used in comparisons of promoter activities. The statistical power of our study was estimated using the genetic power calculator, Quanto v1.2.4 (http://hydra.usc.edu/gxe/) and assuming an additive model of inheritance.
Results
Study Participants’ Demographic Characteristics
The flow diagram of patient selection in the present study is illustrated in Figure E1. A total of 840 subjects, including 224 patients with ARDS and 616 at-risk control subjects were analyzed. Patients who developed ARDS in this population showed higher APACHE III scores than those who did not develop ARDS. The proportion of male subjects or subjects with sepsis, septic shock, pneumonia, and aspiration at ICU admission was also higher in the ARDS group. The distribution of the baseline characteristics of the study population and the comparisons of comorbidities and clinical risk factors between groups are shown in Table 1.
Table 1.
Characteristics of the Study Population: Harborview Medical Center (Seattle, WA) Systemic Inflammatory Response Syndrome Cohort
| At-Risk Control Subjects | ARDS (Mild to Severe) | P Value | |
|---|---|---|---|
| Subjects, n | 616 | 224 | |
| Age, yr | 54.22 ± 16.3 | 55.76 ± 15.13 | 0.2164 |
| Male, n | 371 (60.23) | 161 (71.88) | 0.0019 |
| APACHE III score | 60.51 ± 23.51 | 78.58 ± 25.74 | <0.001 |
| ARDS (moderate and severe) | 0 | 124 (55.35) | <0.001 |
| Risk factor | |||
| Sepsis | 377 (61.20) | 172 (76.79) | <0.001 |
| Sepsis shock | 177 (28.73) | 136 (60.71) | <0.001 |
| Pneumonia | 97 (15.75) | 88 (39.29) | <0.001 |
| Aspiration | 45 (7.31) | 52 (21.88) | <0.001 |
Definition of abbreviations: APACHE, Acute Physiology and Chronic Health Evaluation; ARDS, acute respiratory distress syndrome.
Data are presented as mean ± SD or n (%).
Association of PI3 Variant rs2664581 with Risk of ARDS
The genotypes distributions of SNP rs2664581 in PI3 gene, summarized in Table 2, conformed to Hardy-Weinberg equilibrium. A significant difference in minor allele frequency (MAF) of rs2664581 between the two groups was found (P = 0.0336). Our population yielded greater than 80% power to detect a minimum relative risk (RR) of 1.5 at an α level of 0.05 for variants with an MAF of 0.2 or greater (Table E2). No strong evidence of stratification has been reported in our study population (27).
Table 2.
Genotype Frequencies in the Study Population for At-Risk Control Subjects and Subjects with Acute Respiratory Distress Syndrome
| Genotype Frequencies |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor Allele Frequency |
Controls | Cases | |||||||||
| SNP | Allele | Location | Position* | All | Controls | Cases | P Value† | Genotype | (%) | (%) | HWE P Value‡ |
| rs2664581 | A > C | Exon 2 | 43,237,936 | 0.178 | 0.168 | 0.212 | 0.0336 | AA | 430 (69.81) | 139 (62.05) | 0.298 |
| AC | 165 (26.79) | 77 (34.38) | |||||||||
| CC | 21 (3.41) | 8 (3.57) | |||||||||
Definition of abbreviations: HWE, Hardy-Weinberg equilibrium; SNP, single-nucleotide polymorphism.
Chromosome position based on National Center for Biotechnology Information Build 36.3.
The probability of the Chi-square test for comparison of allele frequencies.
Chi-square test for deviation from HWE in the control group.
Table 3 summarizes the results of the association test between SNP rs2664581 in PI3 and development of ARDS. Assuming dominant and additive models of inheritance, and after adjustment by age, sex, and APACHE III score, we observed a significant association between the minor allele of rs2664581 (C allele) and an increased risk of ARDS (OR = 1.50, 95% CI = 1.07–2.12, P = 0.0198 and OR = 1.35; 95% CI = 1.01–1.80, P = 0.0426, for the dominant and additive models, respectively). The direction and magnitude of association was the same as previously observed (21). Stratified analyses showed a stronger effect of PI3 variants on ARDS susceptibility among the subjects with sepsis (OR = 1.51, 95% CI = 1.04–2.18, P = 0.0297 and OR = 1.44, 95% CI = 1.04–1.99, P = 0.0276, for the dominant and additive models, respectively; Table 3). SNP rs2664581, previously associated with moderate (arterial oxygen pressure [PaO2]/fraction of inspired oxygen [FiO2] ≤ 200 mm Hg but > 100 mm Hg and with PEEP ≥ 5 cm H2O) to severe ARDS (PaO2/FiO2 ≤ 100 mm Hg and with PEEP ≥ 5 cm H2O) accordingly to Berlin criteria (28) (or simply ARDS accordingly to the pre-Berlin criteria) (29), was validated in the current study using a cohort including mild to severe ARDS (PaO2/FiO2 ≤ 300 mm Hg with PEEP ≥ 5 cm H2O, formerly ALI). We performed a sensitivity analysis of the results to test whether differences in the clinical phenotype of mild ARDS versus moderate to severe ARDS (ALI versus ARDS in the pre-Berlin criteria) might influence our findings. The association analyses were repeated after limiting ARDS cases to those with moderate to severe ARDS (n = 124) and using only control subjects without hypoxemic respiratory failure (n = 300). The association between SNP rs2664581 and risk of moderate to severe ARDS was not replicated in the HMC SIRS cohort, although the direction of the effect was in agreement with earlier results (OR = 1.32, 95% CI = 0.80–2.20, P = 0.2780, and OR = 1.17, 95% CI = 0.74–1.85, P = 0.4906, for the dominant and additive models, respectively).
Table 3.
Association between rs2664581 Genotype and Acute Respiratory Distress Syndrome Susceptibility
| All Subjects |
Sepsis |
Nonsepsis |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (n = 840) |
(n = 627) |
(n = 213) |
||||||||||
| Model | Crude OR (95% CI) | P Value | OR adj‡ (95% CI) | P Value | Crude OR (95% CI) | P Value | OR adj‡ (95% CI) | P Value | Crude OR (95% CI) | P Value | OR adj‡ (95% CI) | P Value |
| Additive* | 1.29 (0.98–1.69) | 0.0642 | 1.35 (1.01–1.80) | 0.0426 | 1.38 (1.02–1.87) | 0.0384 | 1.44 (1.04–1.99) | 0.0276 | 0.90 (0.38–2.10) | 0.8016 | 0.944 (0.40–2.21) | 0.8944 |
| Dominant† | 1.41 (1.03–1.95) | 0.0340 | 1.50 (1.07–2.12) | 0.0198 | 1.42 (1.00–2.02) | 0.0473 | 1.51 (1.04–2.18) | 0.0297 | 1.211 (0.40–3.06) | 0.8413 | 1.22 (0.43–3.42) | 0.7067 |
Definition of abbreviations: CI, confidence interval; OR, odds ratio.
In the additive model, ORs were expressed per difference in the number of minor alleles (C allele of rs2664581).
In the dominant model, ORs were expressed as heterozygotes and rare homozygotes compared with common homozygotes (AC/CC versus AA).
Adjusted by age, sex, and Acute Physiology and Chronic Health Evaluation III score.
Functional Characterization of SNPs on PI3 Promoter
Our previous study revealed an association between the coding SNP rs2664581 and altered plasma pre-elafin levels (21). As a result of the high LD between rs2664581 and the remaining SNPs in PI3 gene, PI3 expression and plasma pre-elafin levels could be influenced by SNPs in the promoter region that modify gene transcription by altering the binding of TFs to the promoter or local chromatin architecture. We first describe our investigation of the effect of SNPs in the promoter on the transcriptional activity of PI3 gene. Next, a bioinformatics approach was conducted to identify SNP effects on PI3 TFBSs. Their functional effect on the binding of TFs to the PI3 promoter was examined by EMSA.
PI3 Gene Expression Response to Cytokines Is Promoter Haplotype Dependent
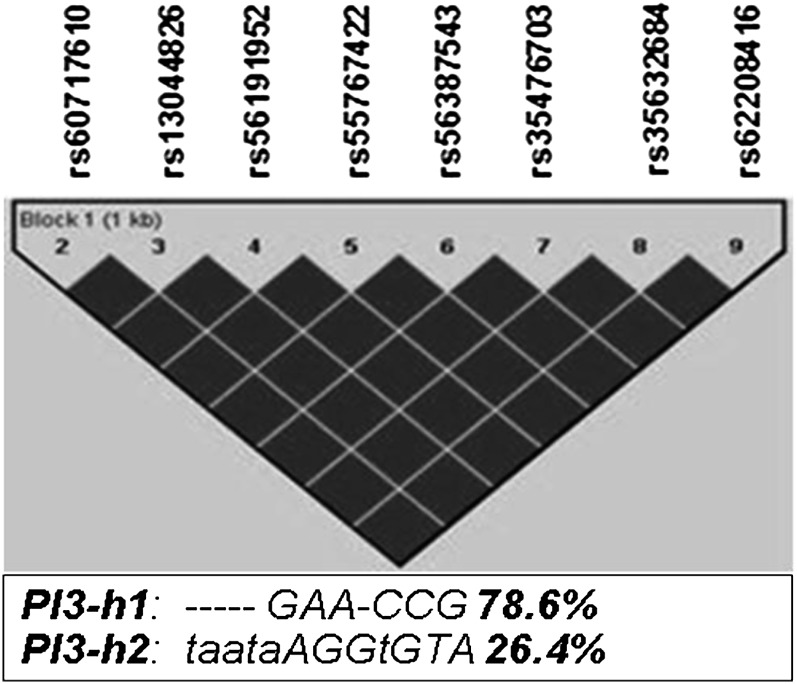
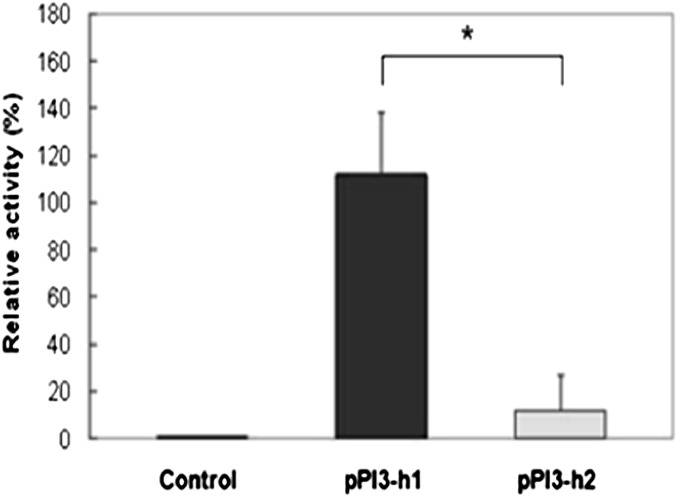
We performed haplotypes analyses using DNA resequencing data from 29 nonrelated, healthy, white subjects, as previously described (21). The pairwise linkage disequilibrium coefficient values showed that all the SNPs in the promoter were in complete LD and collectively segregate into two predominant haplotypes, including a major haplotype (PI3-h1, containing the major alleles of each SNP and with a frequency of 78.6%), and a minor haplotype (PI3-h2, with a frequency of 21.4% consisting of all variant alleles) (Figure 1). IL-1β and TNF-α have been shown to induce significant expression of PI3 in pulmonary epithelial cells (11, 12), suggesting that alveolar epithelial cells may respond to cytokines secreted during the onset of inflammation by increasing their antiproteinase shield. To directly determine an allele-specific effect of the identified genetic variants on cytokine-induced promoter activity of PI3 gene, luciferase reporter constructs were generated using the two PI3 promoter haplotype sequences (PI3-h1 and PI3-h2; Figure E2) and transfected into the A549 alveolar epithelial cells. To optimize the induction of luciferase expression by the A549 cell line in response to cytokines, cells transfected with pPI3-h1 reporter plasmid were stimulated with two different concentrations of IL-1β (20 ng/ml or 100 ng/ml) for 8, 16, 24, and 48 hours, and luciferase assays were then performed. Maximum induction was reached after 24 hours of stimulation with 20 ng/ml of IL-1β (Figure E3A). Under these conditions, we next tested the expression of luciferase by the pPI3-h1 and pPI3-h2 constructs in response to IL-1β. Cells transfected with pPI3-h1 or pPI3-h2 were incubated in the presence of IL-1β. Cells transfected with either pPI3-h1 or pPI3-h2 and incubated in the absence of IL-1β were used as control in our experiment. We confirmed that both constructs were able to respond to cytokine-mediated induction (Figure E3B). The luciferase expression in response to IL-1β was then compared between cells transfected with pPI3-h1 or with pPI3-h2, using cells transfected with empty pEZX-MT01 vector (without PI3 promoter) as a control. Reporter activity was reduced to background levels in cells transfected with pPI3-h2 containing the variant alleles of polymorphisms in PI3 promoter region (Figure 2) when compared with cells transfected with pPI3-h1. A reduction in the transcriptional activity of cells transfected with PI3-h2 was also observed when luciferase activity was induced using TNF-α instead of IL-1β (data not shown).
Figure 1.
Linkage disequilibrium (LD) plot for single-nucleotide polymorphisms (SNPs) in peptidase inhibitor 3 (PI3) promoter region. Linkage disequilibrium coefficient (D′) values between pairs of SNPs were estimated using Haploview based on resequencing data from 29 healthy control subjects (21). Only polymorphisms with a minor allele frequency (MAF) greater than 10% were included in the analysis. Genotypes of SNP PI31 (21) could not be determined by sequencing in over 50% of the subjects, so it was excluded from the LD block determination. LD values were represented as a black-to-white gradient (black, high; white, low). SNPs in the promoter were in complete LD (D’ = 100, black diamonds), and only two predominant haplotypes, PI3-h1 (‐‐‐‐‐GAA-CCG) and PI3-h2 (taataAGGtGTA) with a frequency of 78.6 and 26.4%, respectively, were identified. Please see Ref. 21 for a more detailed description of the alleles at each position.
Figure 2.
Activity of PI3-h1 and PI3-h2 promoters. A549 cells were transfected with pPI3-h1, pPI3-h2, or empty pEZX-MT01 (control) reporter plasmids. At 24 hours after transfection, cells were stimulated with IL-1β (20 ng/ml). The luciferase assay results are presented as relative activity compared with the control (empty pEZX-MT01 vector) as means (± SD) (n = 6). Fold increase was calculated after defining the activity of the empty pEZX-MT01 vector as 1. One-way ANOVA was used in comparisons. *P < 0.05. The differences between cells transfected with pPI3-h1 or pPI3-h2, as well as between the control cells and cells transfected with pPI3-h1 or pPI3-h2, were significant.
Genetic Variants in the PI3 Promoter Influence the Binding of TFs to Core Regulatory Regions
TFs predicted to bind the minor and major alleles of polymorphisms in PI3 promoter are listed in Table E1. Whereas a total of 14 TFBSs are predicted to be haplotype specific for PI3-h2, only 12 are predicted to bind the promoter region defined by PI3-h1. These allele-specific TFBSs may contribute to the marked differences in luciferase activity driven by the pPI3-h1 and pPI3-h2 promoter constructs. The TF NF-κB1 has been proved to modulate the cytokine-mediated induction of human pre-elafin in airway epithelial cells (35). Our in silico analysis predicted an NF-κB1 binding site consensus sequence between −339 and −330 bp. The SNP at −338 (−338G > A), is predicted to abolish this NF-κB1 binding site (Table E1).
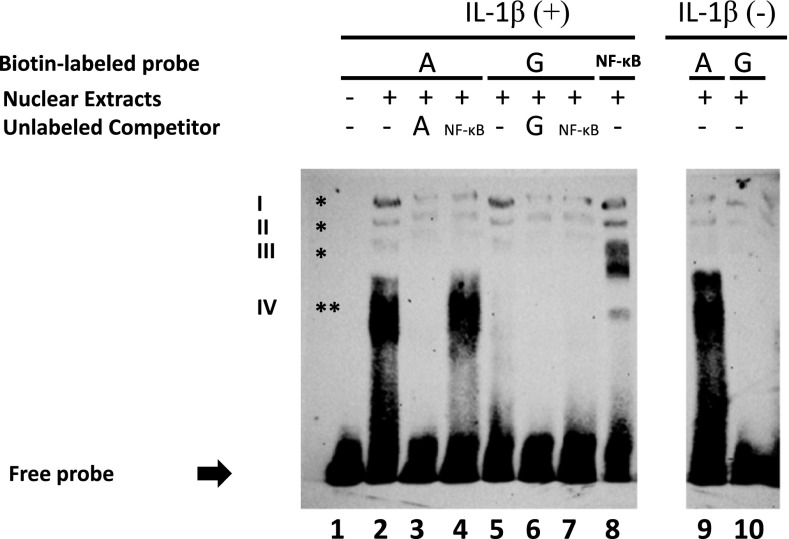
We performed EMSAs to determine whether TF(s) binding differed for the G and A alleles of SNP −338G > A. Incubation of nuclear extracts from 24-hour IL-1β–treated A549 cells with biotin-labeled oligonucleotides containing the G or A alleles (oligos G or A) detected three DNA–protein complexes (I–III; Figure 3, lanes 2 and 5, indicated by an asterisk). Shifted bands (I–III) were completely abolished by an excess of unlabeled G or A oligos (Figure 3, lanes 3 and 6), indicating specific binding of nuclear proteins to the oligonucleotides containing the G or A allele. The same complexes (I–III) were identified when the extracts were incubated with a probe containing NF-κB consensus sequences (oligo NF-κB, lane 8). Complexes I–III identified with oligos G or A were also prevented with an excess of cold oligo NF-κB (lanes 4 and 7), which indicates that the identified complexes probably represent homo- and heterodimers of NF-κB subunits. The intensity of the bands in complexes I–III was significantly reduced when oligo A or G was incubated with NE from untreated cells (lanes 9 and 10), demonstrating that proteins in complexes I–III mainly bind to PI3 promoter in response to cytokine-mediated signaling. A fourth lower complex (IV; Figure 3) was identified only by incubation of nuclear extracts with oligo A, but not with oligo G (lanes 2 and 5, indicated by double asterisks). This complex was completely abolished with an excess of unlabeled oligo A (Figure 3, lane 3), but not when cold oligo NF-κB was used as a competitor (not even at a fold concentration of 500×; Figure 3, lane 4). Complex IV was also observed when nuclear extracts from untreated cells were incubated with oligo A (lane 9).
Figure 3.
Allele-specific effect of SNP −338 (G > A) on binding of nuclear proteins to the promoter of the PI3 gene. Electrophoretic mobility shift assay (EMSA) was performed by mixing nuclear extracts from A549 lung epithelial cells with biotin-labeled oligonucleotides containing the major (G) and minor (A) alleles of SNP −338 (oligo G and A, respectively). Lane 1 corresponds to the free probe (incubation in the absence of nuclear extract). Three different complexes (I–III) were observed when nuclear extracts from cells treated with IL-1β were incubated with either oligo A or G (lane 2 and 5, indicated by single asterisk). Complexes I–III were prevented when unlabeled oligo A (lane 3) or oligo G (lane 6) were added to the binding reaction as cold competitor. The same complexes (I–III) were detected when nuclear extracts was incubated with an oligonucleotide that carried the NF-κB consensus sequence (oligo NF-κB, lane 8). Complexes detected with oligo G or oligo A were also inhibited after the addition of unlabeled oligo NF-κB to the binding reaction (lanes 4 and 7). The intensity of the bands was significantly reduced when oligo A or G were incubated with nuclear extracts from untreated cells (lanes 9 and 10). A fourth complex that migrated lower than complexes I–III was observed only when nuclear extracts from cells treated with IL-1β were incubated with oligo A, but not with oligo G (lanes 2 and 5, indicated by double asterisks). The shifted band (IV) was completely abolished when oligo A was added as a cold competitor (lane 3), but not in the presence of unlabeled oligo NF-κB (lane 4). This fourth complex was also detected in the absence of cytokine induction (lane 9). The image is representative of three independent experiments.
Functional Analysis of rs2664581 Coding Polymorphism
SNP rs2664581, located at exon 2 of PI3 gene, is a nonsynonymous SNP associated with a Threonine–Proline change at residue 34 of the pre-elafin. Comparison of pre-elafin sequences (95 aa) among different species of primates showed that this residue is highly conserved, suggesting a likely functional conservation (Figure E4A). SNP rs2664581 is located in the N-terminal noninhibitory or cementoin domain (residues 1–38) that anchors the molecule to ECM proteins by TGase-catalyzed cross-linking (22–25, 33). AGADIR software was used to predict the effect of rs2664581 in the secondary structure of the cementoin domain of WT and MT pre-elafin proteins. SNP rs2664581 results in a 50% reduction of the helix content in MT protein when compared with WT protein (Figure E4C). Residues 39–95 at the C terminus of pre-elafin represent the noninhibitory domain, a whey acidic protein/four-disulphide core domain with antiproteinase properties (Figure E4B) (18, 36). To investigate the effect of SNP rs2664581 on pre-elafin inhibitory functions and on its ability to bind to ECM proteins, we designed a pre-elafin protein variant (MT), in which the amino acid, Threonine, at position 34, was replaced with Proline (T→P) mimicking the amino acid change associated with SNP rs2664581.
Effect of SNP rs2664581 on the Antiproteinase Activity of Pre-Elafin
SNP rs2664581 is located at the noninhibitory domain of the protein, and, therefore, it is unlikely that it affects the antiproteinase activity of pre-elafin. The serine proteinase–inhibitory activities of the recombinant pre-elafin proteins (WT and MT) were compared by preincubating purified HNE (50 nM) with or without 50 pM-100 nM WT or MT pre-elafin and then measuring residual HNE activity using a fluorogenic substrate that is specific for HNE. Our results showed that WT and MT pre-elafin were both functionally active against HNE, and the half-maximal inhibitory concentrations for WT versus MT recombinant proteins against HNE were similar (Table 4).
Table 4.
Inhibitory Activities of Wild-Type and Mutant Recombinant Pre-Elafin Proteins
| Pre-Elafin Protein | IC50 (50 nM HNE) |
|---|---|
| WT pre-elafin | 5,634 ± 2,571 |
| MT pre-elafin (T34P) | 7,785 ± 1,329 |
Definition of abbreviations: HNE, human neutrophil elastase; IC50, half-maximal inhibitory concentration; MT, mutant; WT, wild-type.
The inhibitory activities of both proteins were compared by preincubating 50 nM HNE in duplicate with or without 50 pM–100 nM WT or MT pre-elafin for 1 hour at 37°C. Residual HNE activity was quantified using a fluorogenic substrate that is specific for HNE and fluorimetry. Dose–response curves were constructed and IC50 of each inhibitor was determined. IC50 values are represented as means (±SD) (n = 3 separate experiments). The IC50 values for WT versus MT pre-elafin were not significantly different.
Effect of SNP rs2664581 on TGase Substrate Properties of Pre-Elafin
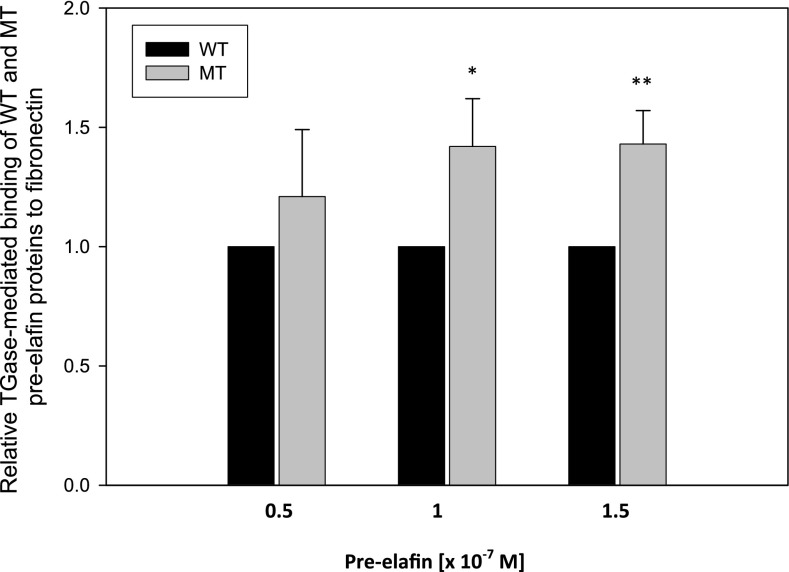
The effect of SNP rs2664581 on the TGase-mediated covalent cross-linking of pre-elafin to ECM proteins was assessed using a published immunoassay method (33). WT and MT pre-elafin recombinant proteins were incubated for 1 hour in the wells of ELISA microplates that had been precoated with fibronectin (to which pre-elafin is known to be cross-linked by TGase [33]) both in the presence and absence of TGase. Pre-elafin that was covalently linked to the immobilized fibronectin was immunodetected using a peroxidase-conjugated antibody system and a chromogenic substrate. We observed a significantly increased binding (P < 0.003) of MT pre-elafin protein to fibronectin when compared with WT pre-elafin (Figure 4). Rabbit polyclonal antibody to human pre-elafin binds to both WT and MT pre-elafin similarly (Figure E5).
Figure 4.
Effect of rs2664581 on transglutaminase (TGase) -mediated binding of wild-type (WT) and mutant (MT) pre-elafin to fibronectin determined by ELISA. WT and MT pre-elafin proteins (0.5, 1, and 1.5 × 10−7 M) were incubated in triplicate wells that had been precoated with fibronectin (1.5 μg/well in 100 μl PBS) for 1 hour at 37°C in the presence and absence of TGase (1.25 × 10−7 M). The wells were washed, and pre-elafin that bound to the immobilized fibronectin was detected by incubating the wells with rabbit polyclonal antibody to human pre-elafin, followed by goat anti-rabbit IgG conjugated to horseradish peroxidase. Peroxidase activity was measured at 450 nm using a chromogenic substrate and H2O2. Results were expressed as relative binding compared with the control (WT protein) after subtracting signals from pre-elafin proteins bound to fibronectin in the absence of TGase. Data are mean (± SD) (n = 3 independent experiments). *WT versus MT (1 × 10−7 M), P < 0.003; **WT versus MT (1.5 × 10–7 M), P < 0.003.
Discussion
In the present study, using an ICU cohort of patients with SIRS at risk for ARDS, we confirmed a previous association between genetic variants in PI3 gene (rs2664581) and ARDS development. The direction and magnitude of association were the same as previously observed, with a stronger effect of PI3 variants on ARDS susceptibility among the subjects with sepsis, supporting previous findings in which the genetic effects of PI3 polymorphisms on ARDS development were modified by the origin of lung injury.
Due to the high LD between the associated SNP rs2664581 and the rest of the SNPs at the PI3 locus, causative variations remain to be identified. We performed a thorough functional characterization of the PI3 promoter SNPs and the coding SNP rs2664581 to determine their effects on PI3 expression and pre-elafin biological functions.
Our data show that polymorphisms in the PI3 promoter affect cytokine-mediated induction of PI3 gene expression in a pulmonary epithelial cell line. Reporter activity in response to cytokines was reduced to background levels in cells transfected with reporter vector pPI3-h2 (containing the minor alleles of polymorphisms in the PI3 promoter region) compared with cells transfected with the pPI3-h1 construct (containing the major alleles of each SNP). Different TFs were predicted to bind the minor and major alleles of polymorphisms in the PI3 promoter (Table E1). These allele-specific TFBSs may contribute to the marked differences that we observed in luciferase activity driven by the pPI3-h1 and pPI3-h2 promoter constructs. NF-κB has been shown to mediate the cytokine-induced activity of the pre-elafin gene promoter (35). This effect is exerted through a functional NF-κB1 site located within 100 bp of the transcription start site (35). This site was not polymorphic in our study. The same study identified a positive-acting region located between −448 and −248 bp (35); however, the binding of NF-κB1 to this region has not been evaluated. Our in silico analysis predicted an NF-κB1 binding site consensus sequence, between −339 and −330 bp, within the previously identified positive-acting region. The SNP at −338 (−338G > A), is predicted to abolish this NF-κB1 binding site. A third NF-κB1 binding site consensus sequence has been previously identified at −972 to −955 (37–39); however, its participation in cytokine-mediated pre-elafin expression has not been tested. Our resequencing analysis revealed a polymorphism (−960 > Del) in this region. This variant is located close to, but not within, the core motif of the NF-κB1 binding site.
We conducted EMSAs to determine the allele-specific effect of SNP −338 (G > A) on binding of TFs to the promoter of PI3 gene. Our results suggest that SNP −338 (G > A) does not affect the binding of NF-κB to PI3 promoter. Complexes I–III detected after incubation of nuclear extracts with an oligonucleotide containing the NF-κB consensus sequence (oligo NF-κB) are also observed after incubation with oligo G or oligo A (Figure 3). Our results also showed a fourth complex detected only in the presence of oligo A, which was not inhibited by addition of unlabeled oligo NF-κB (Figure 3). It is possible to think that the minor allele (A) of SNP −338 may be generating a binding site for the protein(s) in complex IV that overlaps the NF-κB binding site in PI3 promoter. Our luciferase assay showed a reduction in the transcriptional activity of the PI3 promoter carrying the minor alleles of SNPs in the promoter (Figure 2). We speculate that the unidentified protein(s) in complex IV might be acting as a repressor that did not affect the binding of NF-κB to the PI3 promoter, but may interfere with the transcriptional activation mediated by NF-κB in response to cytokines. Additional studies are needed to determine the identity of protein(s) in complex IV.
The observed reduction in the cytokine-induced PI3 gene expression associated with the presence of minor alleles of SNPs in the promoter is in agreement with our previous expression analysis (20), which showed that PI3 gene expression is down-regulated during the acute stage of ARDS. Gene expression levels were well correlated with plasma pre-elafin levels, suggesting a protective role for pre-elafin in ARDS (20). In contrast with these results, we found that the minor allele of the SNP rs2664581 associated with ARDS, and in high LD with minor alleles of SNPs in PI3 promoter, was also associated with increased plasma pre-elafin levels (21). A possible explanation for this apparent contradiction is that the amino acid substitution (T34P) associated with the C allele of SNP rs2664581 results in a defective protein, and a higher level of defective PI3 protein could not provide enough protection against deleterious effects of excessive HNE. To investigate this hypothesis, we examined the functional consequences of SNP rs2664581 on pre-elafin biological functions. SNP rs2664581 is associated with an amino acid change (T34P) at the N terminus of the pre-elafin. The N terminus of the protein or cementoin domain is a sequence of 38 residues containing 5 repeated TGase substrate motifs conforming to the consensus sequence, GQDPVK (Figure E4B). These motifs anchor the molecule to ECM proteins via TGase-catalyzed cross-linking (22–25). TGase-mediated cross-linking of pre-elafin to ECM proteins, including fibronectin, has been reported in a number of in vivo and in vitro studies (22–25, 33, 40, 41). Thus, we investigated the effect of rs2664581 on the capacity of pre-elafin to be cross-linked to ECM proteins by comparing the capacity of WT and MT (T34P) recombinant pre-elafin proteins to undergo TGase-mediated covalent cross-linking to fibronectin in vitro. We observed an increase of up to 50% in the susceptibility of MT pre-elafin protein to be cross-linked to fibronectin by TGase when compared with WT protein (Figure 3). MT pre-elafin bound to fibronectin retained its ability to inhibit NE activity (Figure E6). Activated polymorphonuclear leukocytes (PMNs) that adhere to and migrate on ECM proteins (including fibronectin) are associated with pericellular proteolytic activity that is mediated in part by serine proteinases released from the PMNs and expressed on the PMN surface (42, 43). Thus, after pre-elafin is secreted by lung epithelial cells, its covalent cross-linking to ECM proteins by the action of TGase may serve to target the proteinase inhibitor to a microenvironment in the lung that would enhance its capacity to protect lung ECM proteins from serine proteinase–mediated tissue injury. Although there is no direct evidence that pre-elafin is cross-linked to lung ECM proteins in vivo, high–molecular weight forms of pre-elafin have been detected in lung tissues, probably generated by TGase conjugation (22, 40). Contrary to what we expected, our results suggest that SNP rs2664581 (T34P) augments the proteinase inhibitor’s capacity to locally protect ECM proteins from PMN-mediated degradation. It is also noteworthy that, in addition to its serine proteinase inhibitory properties, pre-elafin has been reported to have opposing activities in regulating the inflammatory response, limiting some inflammatory responses, but increasing others (44–46). Thus, it is possible that the increased TGase-mediated binding of MT pre-elafin to ECM proteins may increase activation of some leukocytes during lung inflammatory responses to injury. Durable binding of MT pre-elafin to ECM proteins may thus result in increased influx and activation of inflammatory cells, leading to lung injury and promoting the development of ARDS. Additional studies are thus needed to compare the proinflammatory properties of WT and MT pre-elafin proteins that have been cross-linked to ECM proteins.
We acknowledge some limitations in our study. We were able to detect a significant association with the development of ARDS, regardless of the severity of the phenotype, especially among subjects with sepsis, with greater than 80% power (Table E2). Inconsistent with earlier results, we did not observe a significant association between rs2664581 polymorphisms and susceptibility to moderate/severe forms of ARDS (although the direction of the effect was consistent with prior reports). This lack of replication may be explained by the small sample size of the moderate/severe subgroups in the HMC cohort. Table E2 shows the statistical power calculated based on the sample size of the moderate/severe ARDS subgroups in the HMC cohort (n = 424). The sample size of this subgroup limits our power to detect all but the strongest effect (RR ≥ 1.6) at an α level of 0.05 for variants with an MAF of 0.2 or greater. Our previous work (21) showed that SNP rs2664581 (MAF = 0.17) was associated with moderate/severe forms of ARDS, with an RR of 1.35. Based on our current sample size, we do not have enough statistical power to detect such a moderate effect. However, for the same reason, our sample size prevents us from discarding a positive association. The lack of replication observed in the current study may not necessarily mean nonexistence of genetic contribution of SNPs in PI3 to the development of moderate/severe ARDS. Larger sample sizes are necessary to discard or confirm this contribution.
We assessed the effect of SNPs in the promoter in the cytokine-induced expression of PI3 gene. In addition to proinflammatory molecules, PI3 expression in lung epithelial is also triggered by HNE (13). The effect of PI3 promoter polymorphisms on the regulation of HNE-mediated pre-elafin expression was not evaluated in our study.
We chose fibronectin as a representative ECM protein on which to compare TGase-mediated cross-linking of WT and MT pre-elafin proteins, as prior studies showed that pre-elafin is efficiently cross-linked by TGase to fibronectin. However, additional studies are needed to assess whether rs2664581 alters TGase-mediated binding of WT and MT recombinant pre-elafin proteins to other ECM components and, if so, whether this associates with development of ARDS. Our data show that SNP rs2664581 has no effect on the inhibitory activity of pre-elafin against HNE. However, pre-elafin also inhibits neutrophil proteinase 3, and the effect of SNP rs2664581 on the inhibitory activity of pre-elafin against proteinase 3 also needs to be determined.
Apart from its well known inhibitory properties, pre-elafin has been found to have broad antimicrobial properties in vitro and in vivo (14, 46–48). Although the biochemical mechanism responsible for the antimicrobial activities has not yet been elucidated, it is well known that the antimicrobial activities of pre-elafin are independent of their antiproteinase function (47–49). Recent studies have demonstrated that the N terminus of pre-elafin, responsible for most of the antimicrobial activity of the protein, adopts an α-helical conformation in the presence of a membrane mimetic, which is typical of many antimicrobial peptides (49). Using AGADIR software, we observed a 50% reduction of the helix content of the cementoin domain of MT pre-elafin in comparison to WT protein (Figure E4C). Additional studies are needed to examine the effect of this change on the antimicrobial properties of MT pre-elafin.
In the current study, functional analyses were limited to all SNPs in the promoter and the coding SNP rs2664581. Besides rs2664581, our resequencing analysis identified two additional nonsynonymous coding SNPs, rs41282752 and rs17333103 (in high LD with rs2664581), which result in two amino acid substitutions (Ala > Thr and Thr > Met) at positions 15 and 17 of the signal peptide sequence of pre-elafin. In addition, five intronic SNPs and six polymorphisms at the 3′ untranslated region, also in high LD with rs2664581, were identified (21). The effect of these genetic variations on the translocation and signal peptide processing, as well as RNA splicing and stability, will require further evaluation.
In summary, our study confirms genetic variants in the PI3 gene as determinants in ARDS susceptibility. However, our results are not conclusive regarding the contribution of SNPs in the PI3 gene to ARDS severity. It is possible that the PI3 gene contributes to the development of ARDS, but not to the severity of the phenotype that may be modulated by a different subset of genes. Studies using larger populations are needed to address this question. Functional analysis of SNPs in PI3 gene showed that genetic variation within the promoter region regulates the cytokine-mediated expression of the PI3 gene. Our results show a differential binding of nuclear proteins to the G or A alleles of SNP −338 in PI3 promoter, which may account for the differential activity observed between PI3-h1 and PI3-h2 promoters. Further investigation beyond the scope of the current study is required to determine the identity of the bound proteins, as well as the mechanism by which these proteins modulate the cytokine-mediated PI3 gene expression and ARDS susceptibility. Coding SNP rs2664581 associated with ARDS risk generates a MT form of pre-elafin with increased ability to bind to ECM components. We speculate that augmented binding of pre-elafin to ECM proteins may enhance the proinflammatory properties of the molecule, and thereby impact ARDS susceptibility. However, our results are inconclusive in regard to the effects of SNP rs2664581 on pre-elafin biological functions and how these functional modifications could promote ARDS development. Future studies using clinically relevant animal models of ALI to compare the ability of WT versus MT pre-elafin in affording protection from ALI could be illuminating regarding the causality of SNP rs2664581 in ARDS development.
Our results point to a potential mechanism by which pre-elafin levels are reduced and/or protein function is altered, thereby resulting in increased susceptibility to ARDS development. Our study provides a rationale for further, detailed, functional characterization of the SNPs in PI3 gene and consideration of using pre-elafin as a novel inhibitor-based anti-inflammatory therapy for ARDS. In addition, the identification of SNPs in PI3 gene relevant to ARDS development will enable us to define a subgroup of patients that may benefit most from pre-elafin supplementation.
Acknowledgments
Acknowledgments
The authors thank Anja Hergrueter and Vanessa Craig for technical assistance, and the staff of the intensive care units at Harborview Medical Center, and all patients and their families for participating in this study.
Footnotes
This work was supported by National Institute of Health grants HL 060710 and ES00002, and by grant CajaMadrid (Spain) (P.T.).
Author Contributions: study concept and design—P.T., D.S.O., C.A.O., J.V., M.W., D.C.C.; acquisition of data—P.T., D.S.O., C.A.O.; analysis and interpretation of data—P.T., D.S.O., C.A.O., J.V., M.W., D.C.C.; drafting of the manuscript—P.T., D.S.O., D.C.C.; critical revision of the manuscript for important intellectual content—P.T., D.S.O., C.A.O., Y.W., Z.W., K.G., L.S., J.V., M.W., D.C.C.; statistical analysis—Y.W., P.T., D.S.O.; obtained funding—M.W., D.C.C.; study supervision—P.T., D.S.O., C.A.O., M.W., D.C.C.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2013-0238OC on March 11, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Lee WL, Downey GP. Leukocyte elastase: physiological functions and role in acute lung injury. Am J Respir Crit Care Med. 2001;164:896–904. doi: 10.1164/ajrccm.164.5.2103040. [DOI] [PubMed] [Google Scholar]
- 2.Belaaouaj A, McCarthy R, Baumann M, Gao Z, Ley TJ, Abraham SN, Shapiro SD. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 3.Kaynar AM, Houghton AM, Lum EH, Pitt BR, Shapiro SD. Neutrophil elastase is needed for neutrophil emigration into lungs in ventilator-induced lung injury. Am J Respir Cell Mol Biol. 2008;39:53–60. doi: 10.1165/rcmb.2007-0315OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawabata K, Hagio T, Matsuoka S. The role of neutrophil elastase in acute lung injury. Eur J Pharmacol. 2002;451:1–10. doi: 10.1016/s0014-2999(02)02182-9. [DOI] [PubMed] [Google Scholar]
- 5.Moraes TJ, Chow CW, Downey GP. Proteases and lung injury. Crit Care Med. 2003;31(4 Suppl):S189–S194. doi: 10.1097/01.CCM.0000057842.90746.1E. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto S, Okayama Y, Shime N, Kimura A, Funakoshi Y, Kawabata K, Ishizaka A, Amaya F. Neutrophil elastase activity in acute lung injury and respiratory distress syndrome. Respirology. 2008;13:581–584. doi: 10.1111/j.1440-1843.2008.01283.x. [DOI] [PubMed] [Google Scholar]
- 7.Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol Rev. 2010;62:726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laskowski M., Jr Protein inhibitors of serine proteinases—mechanism and classification. Adv Exp Med Biol. 1986;199:1–17. doi: 10.1007/978-1-4757-0022-0_1. [DOI] [PubMed] [Google Scholar]
- 9.Sallenave JM, Silva A. Characterization and gene sequence of the precursor of elafin, an elastase-specific inhibitor in bronchial secretions. Am J Respir Cell Mol Biol. 1993;8:439–445. doi: 10.1165/ajrcmb/8.4.439. [DOI] [PubMed] [Google Scholar]
- 10.Saheki T, Ito F, Hagiwara H, Saito Y, Kuroki J, Tachibana S, Hirose S. Primary structure of the human elafin precursor preproelafin deduced from the nucleotide sequence of its gene and the presence of unique repetitive sequences in the prosegment. Biochem Biophys Res Commun. 1992;185:240–245. doi: 10.1016/s0006-291x(05)80981-7. [DOI] [PubMed] [Google Scholar]
- 11.Sallenave JM, Shulmann J, Crossley J, Jordana M, Gauldie J. Regulation of secretory leukocyte proteinase inhibitor (SLPI) and elastase-specific inhibitor (ESI/elafin) in human airway epithelial cells by cytokines and neutrophilic enzymes. Am J Respir Cell Mol Biol. 1994;11:733–741. doi: 10.1165/ajrcmb.11.6.7946401. [DOI] [PubMed] [Google Scholar]
- 12.Pfundt R, Wingens M, Bergers M, Zweers M, Frenken M, Schalkwijk J. TNF-alpha and serum induce SKALP/elafin gene expression in human keratinocytes by a p38 MAP kinase–dependent pathway. Arch Dermatol Res. 2000;292:180–187. doi: 10.1007/s004030050475. [DOI] [PubMed] [Google Scholar]
- 13.Reid PT, Marsden ME, Cunningham GA, Haslett C, Sallenave JM. Human neutrophil elastase regulates the expression and secretion of elafin (elastase-specific inhibitor) in type II alveolar epithelial cells. FEBS Lett. 1999;457:33–37. doi: 10.1016/s0014-5793(99)01004-2. [DOI] [PubMed] [Google Scholar]
- 14.Simpson AJ, Wallace WA, Marsden ME, Govan JR, Porteous DJ, Haslett C, Sallenave JM. Adenoviral augmentation of elafin protects the lung against acute injury mediated by activated neutrophils and bacterial infection. J Immunol. 2001;167:1778–1786. doi: 10.4049/jimmunol.167.3.1778. [DOI] [PubMed] [Google Scholar]
- 15.Tremblay GM, Vachon E, Larouche C, Bourbonnais Y. Inhibition of human neutrophil elastase–induced acute lung injury in hamsters by recombinant human pre-elafin (trappin-2) Chest. 2002;121:582–588. doi: 10.1378/chest.121.2.582. [DOI] [PubMed] [Google Scholar]
- 16.Vachon E, Bourbonnais Y, Bingle CD, Rowe SJ, Janelle MF, Tremblay GM. Anti-inflammatory effect of pre-elafin in lipopolysaccharide-induced acute lung inflammation. Biol Chem. 2002;383:1249–1256. doi: 10.1515/BC.2002.138. [DOI] [PubMed] [Google Scholar]
- 17.Williams SE, Brown TI, Roghanian A, Sallenave JM. SLPI and elafin: one glove, many fingers. Clin Sci (Lond) 2006;110:21–35. doi: 10.1042/CS20050115. [DOI] [PubMed] [Google Scholar]
- 18.Moreau T, Baranger K, Dade S, Dallet-Choisy S, Guyot N, Zani ML. Multifaceted roles of human elafin and secretory leukocyte proteinase inhibitor (SLPI), two serine protease inhibitors of the chelonianin family. Biochimie. 2008;90:284–295. doi: 10.1016/j.biochi.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Sallenave JM. Secretory leukocyte protease inhibitor and elafin/trappin-2: versatile mucosal antimicrobials and regulators of immunity. Am J Respir Cell Mol Biol. 2010;42:635–643. doi: 10.1165/rcmb.2010-0095RT. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Beach D, Su L, Zhai R, Christiani DC. A genome-wide expression analysis in blood identifies pre-elafin as a biomarker in ARDS. Am J Respir Cell Mol Biol. 2008;38:724–732. doi: 10.1165/rcmb.2007-0354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tejera P, Wang Z, Zhai R, Su L, Sheu CC, Taylor DM, Chen F, Gong MN, Thompson BT, Christiani DC. Genetic polymorphisms of peptidase inhibitor 3 (elafin) are associated with acute respiratory distress syndrome. Am J Respir Cell Mol Biol. 2009;41:696–704. doi: 10.1165/rcmb.2008-0410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nara K, Ito S, Ito T, Suzuki Y, Ghoneim MA, Tachibana S, Hirose S. Elastase inhibitor elafin is a new type of proteinase inhibitor which has a transglutaminase-mediated anchoring sequence termed “cementoin”. J Biochem. 1994;115:441–448. doi: 10.1093/oxfordjournals.jbchem.a124357. [DOI] [PubMed] [Google Scholar]
- 23.Steinert PM, Marekov LN. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J Biol Chem. 1995;270:17702–17711. doi: 10.1074/jbc.270.30.17702. [DOI] [PubMed] [Google Scholar]
- 24.Nakane H, Ishida-Yamamoto A, Takahashi H, Iizuka H. Elafin, a secretory protein, is cross-linked into the cornified cell envelopes from the inside of psoriatic keratinocytes. J Invest Dermatol. 2002;119:50–55. doi: 10.1046/j.1523-1747.2002.01803.x. [DOI] [PubMed] [Google Scholar]
- 25.Zeeuwen PL, Hendriks W, de Jong WW, Schalkwijk J. Identification and sequence analysis of two new members of the SKALP/elafin and SPAI-2 gene family: biochemical properties of the transglutaminase substrate motif and suggestions for a new nomenclature. J Biol Chem. 1997;272:20471–20478. doi: 10.1074/jbc.272.33.20471. [DOI] [PubMed] [Google Scholar]
- 26.Glavan BJ, Holden TD, Goss CH, Black RA, Neff MJ, Nathens AB, Martin TR, Wurfel MM. Genetic variation in the FAS gene and associations with acute lung injury. Am J Respir Crit Care Med. 2011;183:356–363. doi: 10.1164/rccm.201003-0351OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Mahony DS, Glavan BJ, Holden TD, Fong C, Black RA, Rona G, Tejera P, Christiani DC, Wurfel MM. Inflammation and immune-related candidate gene associations with acute lung injury susceptibility and severity: a validation study. PLoS One. 2012;7:e51104. doi: 10.1371/journal.pone.0051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, Camporota L, Slutsky AS. Acute respiratory distress syndrome: the Berlin definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 29.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American–European Consensus Conference on ARDS: definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149:818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 32.Owen CA, Campbell MA, Boukedes SS, Campbell EJ. Cytokines regulate membrane-bound leukocyte elastase on neutrophils: a novel mechanism for effector activity. Am J Physiol. 1997;272:L385–L393. doi: 10.1152/ajplung.1997.272.3.L385. [DOI] [PubMed] [Google Scholar]
- 33.Guyot N, Zani ML, Maurel MC, Dallet-Choisy S, Moreau T. Elafin and its precursor trappin-2 still inhibit neutrophil serine proteinases when they are covalently bound to extracellular matrix proteins by tissue transglutaminase. Biochemistry. 2005;44:15610–15618. doi: 10.1021/bi051418i. [DOI] [PubMed] [Google Scholar]
- 34.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 35.Bingle L, Tetley TD, Bingle CD. Cytokine-mediated induction of the human elafin gene in pulmonary epithelial cells is regulated by nuclear factor-κB. Am J Respir Cell Mol Biol. 2001;25:84–91. doi: 10.1165/ajrcmb.25.1.4341. [DOI] [PubMed] [Google Scholar]
- 36.Schalkwijk J, Wiedow O, Hirose S. The trappin gene family: proteins defined by an N-terminal transglutaminase substrate domain and a C-terminal four-disulphide core. Biochem J. 1999;340(Pt 3):569–577. [PMC free article] [PubMed] [Google Scholar]
- 37.King AE, Morgan K, Sallenave JM, Kelly RW. Differential regulation of secretory leukocyte protease inhibitor and elafin by progesterone. Biochem Biophys Res Commun. 2003;310:594–599. doi: 10.1016/j.bbrc.2003.08.151. [DOI] [PubMed] [Google Scholar]
- 38.Pol A, Pfundt R, Zeeuwen P, Molhuizen H, Schalkwijk J. Transcriptional regulation of the elafin gene in human keratinocytes. J Invest Dermatol. 2003;120:301–307. doi: 10.1046/j.1523-1747.2003.12043.x. [DOI] [PubMed] [Google Scholar]
- 39.Chowdhury MA, Kuivaniemi H, Romero R, Edwin S, Chaiworapongsa T, Tromp G. Identification of novel functional sequence variants in the gene for peptidase inhibitor 3. BMC Med Genet. 2006;7:49. doi: 10.1186/1471-2350-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molhuizen HO, Alkemade HA, Zeeuwen PL, de Jongh GJ, Wieringa B, Schalkwijk J. SKALP/elafin: an elastase inhibitor from cultured human keratinocytes: purification, cDNA sequence, and evidence for transglutaminase cross-linking. J Biol Chem. 1993;268:12028–12032. [PubMed] [Google Scholar]
- 41.Muto J, Kuroda K, Wachi H, Hirose S, Tajima S. Accumulation of elafin in actinic elastosis of sun-damaged skin: elafin binds to elastin and prevents elastolytic degradation. J Invest Dermatol. 2007;127:1358–1366. doi: 10.1038/sj.jid.5700647. [DOI] [PubMed] [Google Scholar]
- 42.Owen CA, Campbell MA, Sannes PL, Boukedes SS, Campbell EJ. Cell surface–bound elastase and cathepsin G on human neutrophils: a novel, non-oxidative mechanism by which neutrophils focus and preserve catalytic activity of serine proteinases. J Cell Biol. 1995;131:775–789. doi: 10.1083/jcb.131.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell EJ, Campbell MA, Boukedes SS, Owen CA. Quantum proteolysis by neutrophils: implications for pulmonary emphysema in alpha 1-antitrypsin deficiency. J Clin Invest. 1999;104:337–344. doi: 10.1172/JCI6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson AJ, Cunningham GA, Porteous DJ, Haslett C, Sallenave JM. Regulation of adenovirus-mediated elafin transgene expression by bacterial lipopolysaccharide. Hum Gene Ther. 2001;12:1395–1406. doi: 10.1089/104303401750298553. [DOI] [PubMed] [Google Scholar]
- 45.Sallenave JM, Cunningham GA, James RM, McLachlan G, Haslett C. Regulation of pulmonary and systemic bacterial lipopolysaccharide responses in transgenic mice expressing human elafin. Infect Immun. 2003;71:3766–3774. doi: 10.1128/IAI.71.7.3766-3774.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMichael JW, Roghanian A, Jiang L, Ramage R, Sallenave JM. The antimicrobial antiproteinase elafin binds to lipopolysaccharide and modulates macrophage responses. Am J Respir Cell Mol Biol. 2005;32:443–452. doi: 10.1165/rcmb.2004-0250OC. [DOI] [PubMed] [Google Scholar]
- 47.Baranger K, Zani ML, Chandenier J, Dallet-Choisy S, Moreau T. The antibacterial and antifungal properties of trappin-2 (pre-elafin) do not depend on its protease inhibitory function. FEBS J. 2008;275:2008–2020. doi: 10.1111/j.1742-4658.2008.06355.x. [DOI] [PubMed] [Google Scholar]
- 48.Bellemare A, Vernoux N, Morisset D, Bourbonnais Y. Human pre-elafin inhibits a Pseudomonas aeruginosa–secreted peptidase and prevents its proliferation in complex media. Antimicrob Agents Chemother. 2008;52:483–490. doi: 10.1128/AAC.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bellemare A, Vernoux N, Morin S, Gagne SM, Bourbonnais Y. Structural and antimicrobial properties of human pre-elafin/trappin-2 and derived peptides against Pseudomonas aeruginosa. BMC Microbiol. 2010;10:253. doi: 10.1186/1471-2180-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]