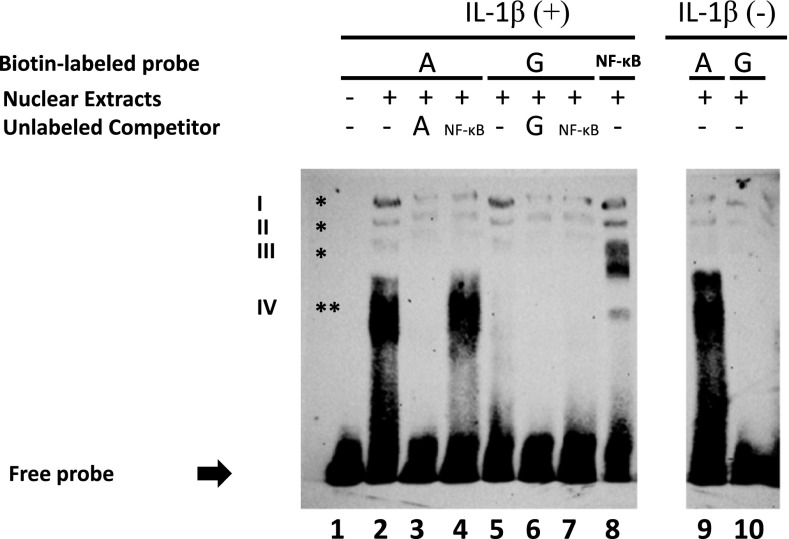
Figure 3.
Allele-specific effect of SNP −338 (G > A) on binding of nuclear proteins to the promoter of the PI3 gene. Electrophoretic mobility shift assay (EMSA) was performed by mixing nuclear extracts from A549 lung epithelial cells with biotin-labeled oligonucleotides containing the major (G) and minor (A) alleles of SNP −338 (oligo G and A, respectively). Lane 1 corresponds to the free probe (incubation in the absence of nuclear extract). Three different complexes (I–III) were observed when nuclear extracts from cells treated with IL-1β were incubated with either oligo A or G (lane 2 and 5, indicated by single asterisk). Complexes I–III were prevented when unlabeled oligo A (lane 3) or oligo G (lane 6) were added to the binding reaction as cold competitor. The same complexes (I–III) were detected when nuclear extracts was incubated with an oligonucleotide that carried the NF-κB consensus sequence (oligo NF-κB, lane 8). Complexes detected with oligo G or oligo A were also inhibited after the addition of unlabeled oligo NF-κB to the binding reaction (lanes 4 and 7). The intensity of the bands was significantly reduced when oligo A or G were incubated with nuclear extracts from untreated cells (lanes 9 and 10). A fourth complex that migrated lower than complexes I–III was observed only when nuclear extracts from cells treated with IL-1β were incubated with oligo A, but not with oligo G (lanes 2 and 5, indicated by double asterisks). The shifted band (IV) was completely abolished when oligo A was added as a cold competitor (lane 3), but not in the presence of unlabeled oligo NF-κB (lane 4). This fourth complex was also detected in the absence of cytokine induction (lane 9). The image is representative of three independent experiments.