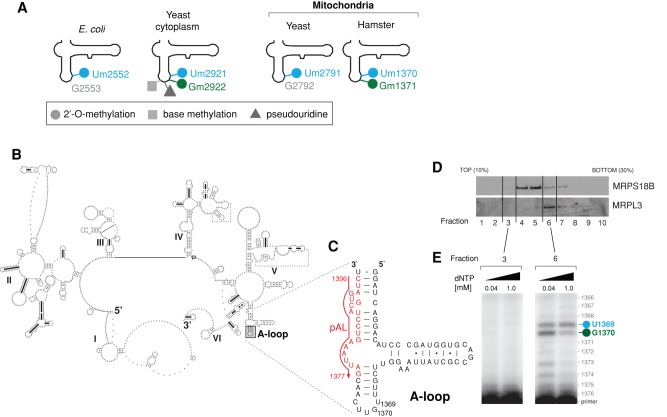
FIGURE 1:
Modification of the 16S rRNA A-loop in human mitochondria. (A) Comparison of the modification status of LSU A-loops across the tree of life. The various modifications reported previously in the LSU rRNA A-loop from E. coli (Caldas et al., 2000b), yeast mitochondria (Pintard et al., 2002a), and hamster (Dubin, 1974; Dubin and Taylor, 1978; Baer and Dubin, 1981) and the yeast cytosol are indicated. (B) The predicted secondary structure of human mitochondrial 16S rRNA. Human mitochondrial 16S rRNA was folded using the thermodynamics-based method of Zuker and Turner (Fields and Gutell, 1996) and downloaded from the Comparative RNA Web (CRW) database (Cannone et al., 2002). The position of the A-loop is indicated by a gray box. (C) Strategy of primer extension used to detect modifications of the A-loop in human mitochondrial 16S rRNA. The sequence encompassing the 16S rRNA A-loop is shown (16S nucleotide positions: 1325–1397). The position of the primer (pAL) used for reverse transcription primer extension (RT-PEx) is indicated in red. (D) Fractionation of the mitochondrial ribosome. Mitochondria that were isolated from HeLa cells were lysed, separated through a 10–30% (wt/vol) isokinetic sucrose gradient, and fractionated. Ten fractions were collected and analyzed by Western blotting using antibodies specific for a component of mt-LSU (MRPL3) and a component of mt-SSU (MRPS18B). (E) Detection of the A-loop modifications in human mitochondrial 16S rRNA. RNA isolated from fractions 3 and 6 of the sucrose gradient from D was subjected to RT-PEx using a radioactively labeled primer as shown in C, using two concentrations of dNTPs as indicated. Samples were analyzed on a 15% denaturing polyacrylamide gel and subjected to autoradiography. The specific pausing sites for 16S rRNA nucleotide positions U1369 (blue) and G1370 (green) are indicated.