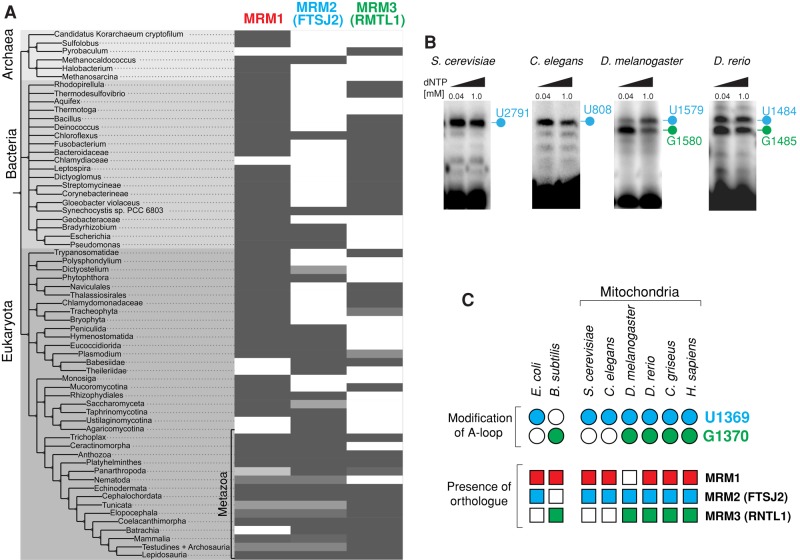
FIGURE 3:
Evolutionary distribution of MTase orthologues and specific modification of the A-loop of LSU rRNA. (A) Evolutionary analysis of human MRM1, MRM2, and MRM3 across higher taxa. The tree shows higher taxa for human MRM1, MRM2, and MRM3 based on National Center for Biotechnology Information taxonomy and reciprocal BLAST hits computed for 147 fully sequenced genomes (Reference Proteomes, 2013_04 release). The percentage of hits within a taxon is shown by a grayscale, with the darker tones representing the higher percentages. Light gray may indicate the presence of evolutionary divergence within the taxon, incomplete proteome data, or limitations of the reciprocal BLAST approach. (B) Detection of A-loop modifications in mitochondrial LSU rRNAs of several eukaryotic species. RNA isolated from S. cerevisiae, C. elegans, D. melanogaster, and D. rerio was subjected to RT-PEx using a radioactively labeled primer binding upstream from the A-loop. Samples were analyzed on a 15% denaturing polyacrylamide gel and subjected to autoradiography. The specific pausing sites for the LSU rRNA A-loop are indicated. Nucleotide positions that correspond to human 16S rRNA U1369 are indicated in blue, whereas those corresponding to human 16S rRNA U1370 are in green. (C) Correlation between the presence of MTase orthologues and specific modifications of the A-loop. The presence of MRM1 (red), MRM2 (blue), or MRM3 (green) in analyzed species is indicated by filled squares. Open squares indicate that an orthologue has not been identified. The modification status of LSU nucleotides corresponding to the human 16S residues U1369 (blue) and G1370 (green) is represented by circles. Open circles indicate a lack of modification; filled circles indicate that the nucleotide is modified. The data for Bacillus subtilis are those of Hansen et al. (2002), and other references can be found in the text.