Abstract
Congenital myasthenic syndromes (CMSs) are heterogeneous neuromuscular disorders characterized by skeletal muscle weakness caused by disruption of signal transmission across the neuromuscular junction (NMJ). CMSs are rarely encountered in veterinary medicine, and causative mutations have only been identified in Old Danish Pointing Dogs and Brahman cattle to date. Herein, we characterize a novel CMS in 2 Labrador Retriever littermates with an early onset of marked generalized muscle weakness. Because the sire and dam share 2 recent common ancestors, CMS is likely the result of recessive alleles inherited identical by descent (IBD). Genome-wide SNP profiles generated from the Illumina HD array for 9 nuclear family members were used to determine genomic inheritance patterns in chromosomal regions encompassing 18 functional candidate genes. SNP haplotypes spanning 3 genes were consistent with autosomal recessive transmission, and microsatellite data showed that only the segment encompassing COLQ was inherited IBD. COLQ encodes the collagenous tail of acetylcholinesterase, the enzyme responsible for termination of signal transduction in the NMJ. Sequences from COLQ revealed a variant in exon 14 (c.1010T>C) that results in the substitution of a conserved amino acid (I337T) within the C-terminal domain. Both affected puppies were homozygous for this variant, and 16 relatives were heterozygous, while 288 unrelated Labrador Retrievers and 112 dogs of other breeds were wild-type. A recent study in which 2 human CMS patients were found to be homozygous for an identical COLQ mutation (c.1010T>C; I337T) provides further evidence that this mutation is pathogenic. This report describes the first COLQ mutation in canine CMS and demonstrates the utility of SNP profiles from nuclear family members for the identification of private mutations.
Introduction
Skeletal muscle contraction is stimulated by the emission of the neurotransmitter acetylcholine (ACh) by the motor neuron, and terminated by acetylcholinesterase (AChE) in the neuromuscular junction (NMJ). Disruption of signal transmission within the NMJ resulting from presynaptic, synaptic, or post-synaptic defects causes congenital myasthenic syndromes (CMSs), heterogeneous neuromuscular disorders characterized by skeletal muscle weakness and fatigue. Mutations causing CMSs in humans have been identified in 18 genes to date, with a majority of cases attributed to CHRNE, COLQ, RAPSN, and DOK7 [1]. Mutations are predominantly autosomal recessive, and often act in compound heterozygosity [2]–[5].
Naturally-occurring CMSs are rarely described in veterinary medicine; when they do occur, they are usually in animals between 6 to 12 weeks of age, appear to be familial, and are characterized by severe generalized skeletal muscle weakness. The first report of canine CMS was in the Jack Russell Terrier in 1974 [6]. Since that time, acetylcholine receptor (AChR) deficiency has been confirmed in Jack Russell Terriers [7], as well as Smooth Fox Terriers [8]. CMS has also been clinically described in families of Springer Spaniels [9], Miniature Smooth-Haired Dachshunds [10], and Old Danish Pointing Dogs [11]. Characterization of CMS at the molecular level has only been achieved in Old Danish Pointing Dogs (missense mutation in CHAT) [12], and in young Brahman cows (deletion in CHRNE) [13].
Diagnosis of CMS is challenging and relies on clinical evaluation, morphological studies of muscle and peripheral nerves, electrodiagnostic studies, the absence of serum antibodies against muscle AChRs, demonstration of AChR deficiency, and most recently, molecular genetic studies. We have identified a novel canine CMS in a family of Labrador Retrievers. Affected littermates exhibited signs clinically distinct from neuromuscular disorders previously characterized in the breed: exercise-induced collapse (EIC) [14], centronuclear myopathy (CNM) [15], and myotubular myopathy [16].
While genome-wide association studies (GWAS) are an efficient approach for the identification of recessive alleles, they require several unrelated affected individuals [17]–[19]. In the absence of a population suitable for GWAS, we utilized genome-wide SNP profiles from a nuclear family to evaluate inheritance patterns in chromosomal regions harboring all 18 candidate genes. Described herein is the clinical characterization of CMS in a Labrador Retriever family and identification of a missense mutation in COLQ.
Materials and Methods
Animals
The dogs evaluated in this study were members of a Labrador Retriever family. The dam and sire were clinically normal and both tested clear for the EIC [14] and CNM [15] mutations affecting the Labrador Retriever breed. At 6 weeks of age, 2 female puppies from a litter of 9 were presented to the Texas A&M University (TAMU) Veterinary Teaching Hospital with a 3- to 4-week history of exercise-induced tetraparesis. One puppy was euthanized for progression of clinical signs at 7 weeks of age. Further evaluation including electrophysiology and blood and tissue collection was performed prior to euthanasia of the second puppy. No clinical signs of weakness were observed in 6 littermates (1 puppy died at birth), and a neuromuscular disease had not been previously identified in this family.
Sample Collection and DNA Isolation
Whole blood was drawn from family members by their primary care veterinarians. Buccal swabs were collected from additional Labrador Retrievers, both related and unrelated to the affected dogs. DNAs from Labrador Retrievers and other breeds previously collected for unrelated studies were also available. Informed owner consent and all samples were obtained in accordance with protocols approved by the Clemson University Institutional Review Board (IBC2008-17). Owner consent was obtained for post-mortem tissue collection from both affected puppies at TAMU. Genomic DNA was isolated using Gentra Puregene Blood Kit (Qiagen).
Electrophysiology
Electrodiagnostics on the affected second puppy, including electromyography (EMG), measurement of motor nerve conduction velocity (MNCV), and measurement of the decrement in the compound muscle action potential (CMAP) following repetitive nerve stimulation, were performed on the left side under general inhalational anesthesia using a Nicolet Viking Select EMG/evoked potential system (Nicolet, Biomedical Inc.). Insulated stainless steel needle electrodes were used for both nerve stimulation and recording from muscle, while a platinum subdermal electrode (Grass-Telefactor) was employed as a ground. MNCV of the peroneal and ulnar nerves was determined by dividing the distance between proximal and distal stimulation sites by the difference in latency of the corresponding CMAP recorded from the extensor digitorum brevis muscle and palmar interosseous muscles, respectively, after supramaximal stimulation (2 Hz stimulus rate, 0.2 ms stimulus duration). Amplitude (peak to peak) was measured from CMAPs derived from stimulation at the proximal and distal stimulation sites. Repetitive nerve stimulation parameters included stimulation frequencies of 1, 2, 3, 5, 10, 20, or 50 Hz.
Histopathology, Histochemistry, and Immunohistochemistry
Specimens collected post-mortem from the second puppy included the infraspinatus, extensor carpi radialis, triceps brachii, biceps femoris, quadriceps, and cranial tibial muscles on the right side. The muscles were frozen in isopentane pre-cooled in liquid nitrogen and stored at –80°C until further processing. Light microscopic evaluation of histological and histochemical stains and reactions was performed according to standard protocols [20] and included hematoxylin and eosin, modified Gomori trichrome, periodic acid Schiff, phosphorylase, esterase, myofibrillar ATPase reaction at preincubation pH of 9.8, 4.5, and 4.3, reduced nicotinamide adenine dinucleotide-tetrazolium reductase, succinic dehydrogenase, acid phosphatase, and oil red O.
Specimens from the radial and peroneal nerves were immersion fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer before shipment to the laboratory. Upon receipt, nerves were post-fixed in 1% aqueous osmium tetroxide for 3 h to 4 h and then dehydrated in a graded alcohol series and propylene oxide. After infiltration with a 1:1 mixture of propylene oxide and araldite resin for 4 h, nerves were placed in 100% araldite resin overnight and then embedded in fresh araldite resin. Thick sections (1 µm) were cut and stained with paraphenylediamine prior to light microscopic evaluation.
For immunohistochemical localization of motor end-plates, serial cryosections (8 µm) were obtained from the external intercostal muscle of 1 affected Labrador Retriever, archived frozen muscle of a previously diagnosed Jack Russell Terrier with CMS due to AChR deficiency (neuromuscular disease control), and a normal dog (wild-type control). Sections from each dog were incubated with the esterase reaction for identification of presumptive motor end-plates or Alexa Fluor 594 α-bungarotoxin (1:1000, Molecular Probes) for localization of AChRs at the motor end-plate. Serial sections were evaluated with light microscopy (esterase reaction) and fluorescent microscopy (red fluorescence) and localization of stainings compared.
AChR Quantification and Antibody-Bound AChR
AChR was extracted from external intercostal muscle specimens of both affected puppies by a procedure modified from that of Lindstrom and Lambert [21]. The muscle specimens were stored at –70°C prior to homogenization and extraction of AChR in 2% Triton X-100. Solubilized AChRs were labeled by incubation with an excess of 125I-α-bungarotoxin (125I-αbgt) followed by sequential addition of high titer rat-anti-AChR antibody and precipitation with goat anti-rat IgG. The precipitate was pelleted, washed, and quantitated in a gamma counter. The amount of AChR complexed with 125I-αbgt was quantified and expressed in terms of moles of 125I-αbgt precipitated per gram of tissue. The concentration of in-situ antibody-AChR complexes was determined by precipitation with goat anti-rat serum in the presence of normal rat serum. Quantitative serum AChR antibody concentrations were determined as previously described using an immunoprecipitation radioimmunoassay procedure [22].
SNP Profiling
Eighteen candidate genes were selected based on their involvement in human CMSs. Candidate genes were distributed across 13 canine chromosomes: 3 (DOK7), 5 (AGRN, CHRNB1, CHRNE, DPAGT1), 6 (ALG14), 9 (SCN4A), 10 (GFPT1), 11 (ALG2, MUSK), 13 (PLEC1), 18 (RAPSN), 20 (LAMB2), 23 (COLQ), 25 (CHRNG, CHRND), 28 (CHAT), and 36 (CHRNA1) [1]. The Illumina CanineHD Infinium BeadChip was used to profile 173,662 SNPs representing all chromosomes for 9 nuclear family members: 7 littermates and both parents (Figure 1). Arrays were processed according to manufacturer’s protocols. Allele frequencies were calculated using a case/control analysis conducted with PLINK [23]. Chromosomal regions harboring candidate genes were evaluated for SNP haplotypes with frequencies of 1.0 in the CMS cases and between 0.14 and 0.50 in the controls. Individual genotypes in regions fitting these parameters were then manually examined to ensure that no unaffected littermates were homozygous for the allele present in the affected dogs.
Figure 1. Multigenerational pedigree of Labrador Retrievers.
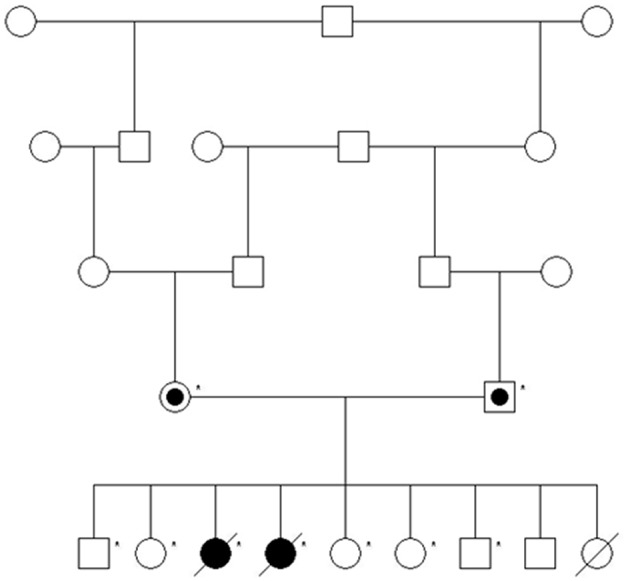
Filled individual icons denote affected dogs and semi-filled icons denote obligate carriers. Asterisks mark individuals selected for whole-genome SNP profiling.
Microsatellite Analysis
PCR for amplification of individual microsatellite markers was performed using materials and thermal cycling parameters previously described [24], [25]. Products were resolved with GeneScan 600 LIZ size standard (Applied Biosystems) on an ABI 3730XL DNA Analyzer (Applied Biosystems). GeneMapper Software version 4.0 (Applied Biosystems) was used to visualize the microsatellite signals.
Sanger Sequencing
Primers were designed to amplify all coding regions and splice sites of CHRNG, CHRND, and COLQ. Products were amplified using ReddyMix Master Mix (Thermo Scientific) with 0.4 µM of each primer and 50 ng of DNA. Products were purified using the Qiax II Gel Extraction Kit (Qiagen) or an ExoSAP master mix consisting of 0.5 units of Exonuclease I (New England BioLabs) and 0.25 units of SAP (Promega). Primer sequences and purification methods for each amplicon are given in Table S1. Sequencing products were resolved on an ABI 3730XL DNA Analyzer (Applied Biosystems).
Restriction Enzyme Digest
Detection of the COLQ variant was accomplished using the endonuclease BtsI, which recognizes the following sequence and cutsite (∧): 5′ …NN∧CACTGC…3′. The digestion was performed using BtsI CutSmart (New England BioLabs) with 2.5 µL 10X buffer, 0.40 µL BtsI, 1 µg DNA, and water adjusted accordingly for a total reaction volume of 25 µL. Products were resolved on a 2% agarose gel.
Results
Clinical Description
Neurological examination was consistent with a generalized neuromuscular disease with marked short-strided tetraparesis that worsened with exercise. Postural reactions were preserved with the exception of hopping which was diminished in all limbs when the puppies were made to bear full weight. Spinal reflexes including the patellar, cranial tibial, and flexor withdrawals were reduced in all limbs. A pyridostigmine bromide challenge resulted in worsening of muscle weakness.
Electrophysiology
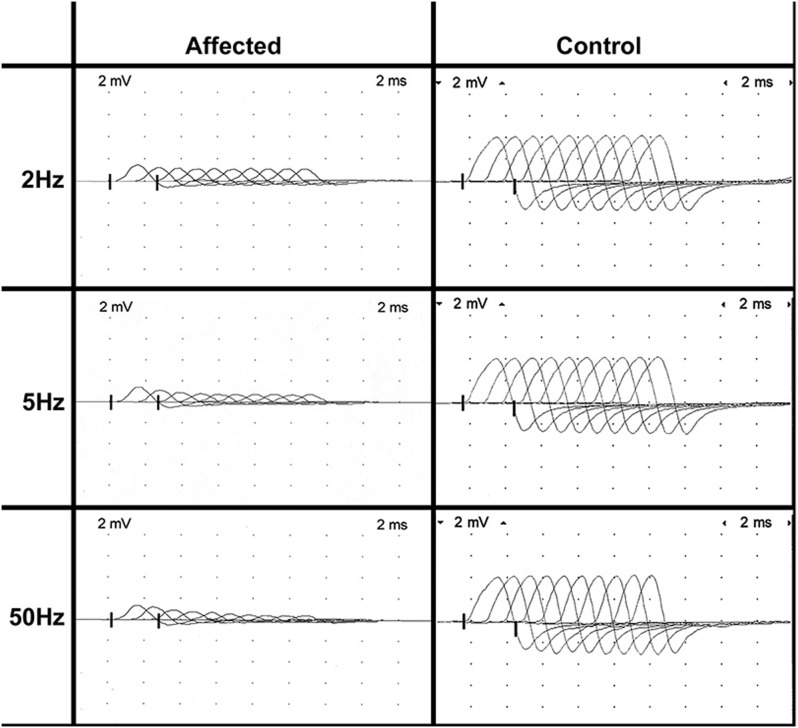
EMG was performed on the left side of the body and included the supraspinatus, infraspinatus, triceps, biceps, extensor carpi radialis, superficial and deep digital flexor, interosseous, biceps femoris, rectus femoris, cranial tibial, and gastrocnemius muscles. Abnormal EMG activity was not identified in any muscle group. Peroneal and ulnar MNCV was likely age appropriate (41 m/sec and 31 m/sec, respectively; reported values for normal dogs 1 to 6 months of age 32 m/sec to 55 m/sec and 25 m/sec to 48 m/sec) [26], [27]. The peroneal and ulnar nerve CMAP amplitudes were reduced at all stimulus sites (1.9 mV to 2.1 mV and 2.6 mV to 2.9 mV, respectively, reference ≥8 mV, peak-to-peak) (Figure 2) [27]. F-waves were not recordable upon stimulation of either the peroneal or ulnar nerve. Repetitive stimulation of the peroneal nerve revealed a decrement in the CMAP amplitude of 22% at 2 Hz, 33% at 5 Hz, and 35% at 50 Hz when the first and third CMAPs were compared (Figure 3). Decrements greater than 10% are considered significant.
Figure 2. Peroneal MNCV of an affected Labrador Retriever recorded at the extensor digitorum brevis muscle with stimulation at the level of the hock, stifle, and hip.
Although the CV was normal considering the dog’s age, the amplitude of the CMAP was uniformly diminished. The timebase between vertical columns on the x-axis is 2 msec and the voltage measured between adjacent rows on the y-axis is 2 mV. Control tracings are from the peroneal nerve of a healthy 5 month old Beagle. Note: labeling varies with the control amplitudes measured from baseline to peak, peak-to-peak amplitudes are within normal limits (≥8 mV). Latency 2 markings also vary.
Figure 3. Repetitive stimulation of the peroneal motor nerve of an affected Labrador Retriever at 2 Hz (A), 5 Hz (B), and 50 Hz (C).
Decrement of the CMAP was observed at all tested frequencies. Sweep speed and sensitivity settings are identical to those in Figure 2. Control tracings are from the peroneal nerve of a healthy 5 month old Beagle with no decrement seen at low frequency stimulation and normal pseudofaciliation (CMAP gets taller and narrower) with tetanic stimulation.
Histopathology, Histochemistry, and Immunohistochemistry
Myofiber size was appropriate in all muscles evaluated and no specific abnormalities were identified, making a congenital myopathy unlikely. Multifocal areas of esterase reactivity were identified, but it could not be determined from this reaction if staining correlated with localization of motor end-plates. In the Labrador Retriever with CMS, esterase staining was evident in multiple locations but did not fully correlate with AChR labeling (Figure 4). There was a good correlation with esterase staining and AChR labeling in the normal dog. In the Jack Russell Terrier with CMS (neuromuscular disease control), several end-plates were stained with esterase; however, no AChRs were labeled in the serial muscle section, consistent with the marked decrease in muscle AChRs described in this breed. There was no evidence of axonal degeneration, demyelination, or abnormalities of supporting structures in the peripheral nerve evaluations, excluding a congenital polyneuropathy.
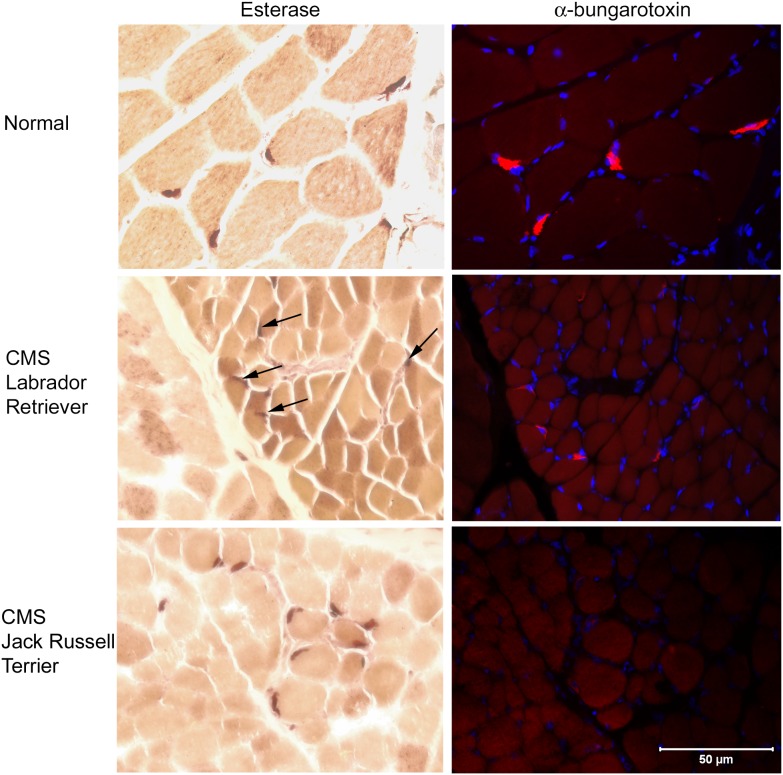
Figure 4. Cryosections (8 µm) of intercostal muscle from a normal dog, a Labrador Retriever with CMS (end-plate AChE deficiency), and a Jack Russell Terrier with CMS due to AChR deficiency (neuromuscular disease control) are illustrated.
For each dog, histochemical staining for esterase activity (brown stain) is shown along with a serial section demonstrating immunofluorescent localization of α-bungarotoxin for AChR and end-plate localization (red color). Muscle nuclei are blue (Dapi stain). There is a good correlation between esterase staining (brown) and α-bungarotoxin localization (red) in the control dog muscle. Although esterase staining is present in the Labrador Retriever muscle (arrows), the localization correlates poorly with that of AChRs. In the CMS Jack Russell Terrier esterase staining was present; however, staining for AChR was markedly decreased or absent, consistent with a markedly decreased AChR content. Bar = 50 µm for all images.
AChR Quantification and Antibody-Bound AChR
The concentration of AChR was determined from external intercostal muscle samples collected origin to insertion following euthanasia of both affected Labrador Retriever puppies. The AChR concentration was decreased in both puppies (0.07 pmol/gm and 0.10 pmol/gm tissue, reference 0.2 pmol/gm to 0.4 pmol/gm). AChR antibodies were not detected bound to muscle AChRs or in the serum.
Analysis of Family Genomic Inheritance Patterns
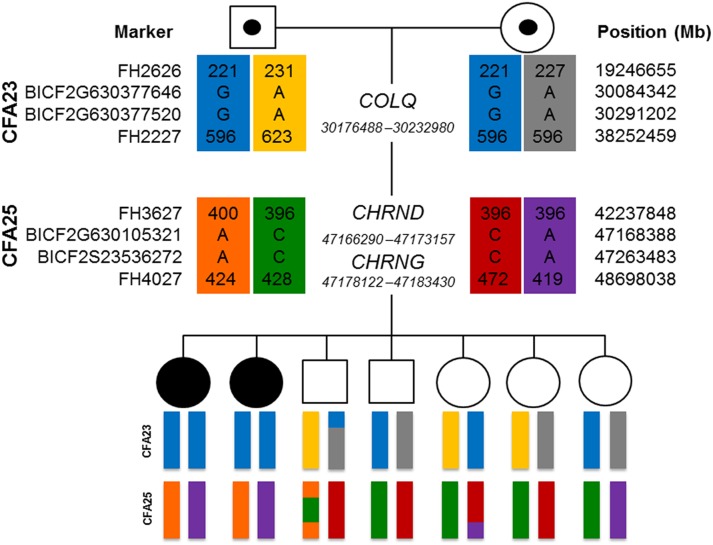
Over 172,000 SNPs across 40 canine chromosomes were genotyped for each of 9 nuclear family members. Allele frequencies in regions on the 13 chromosomes harboring candidate genes were evaluated for a pattern consistent with a recessive trait (Table S2). SNP haplotypes on both chromosomes 23 and 25 were homozygous in the cases and had frequencies of 0.36 and 0.29, respectively, in the unaffected individuals. Chromosome 23 harbors COLQ, while the region on chromosome 25 includes both CHRNG and CHRND.
Because the sire and dam share 2 recent common ancestors, we hypothesized that the causative mutation was inherited identical by descent (IBD). To determine whether the aforementioned segments of chromosomes 23 and 25 were inherited IBD, we genotyped polymorphic microsatellite markers in each region. Genotypes revealed homozygosity in the affected dogs for markers flanking COLQ, providing evidence that the segment on chromosome 23 is IBD (Figure 5). Affected dogs were heterozygous for haplotypes encompassing CHRNG and CHRND, suggesting that the chromosome 25 segment was not inherited from a recent common ancestor.
Figure 5. Microsatellite and SNP haplotypes (color-coded bars below individuals) are shown for 3 candidate genes.
Positions (in Mb) are according to CanFam 2. Filled individual icons denote affected dogs and semi-filled icons denote obligate carriers. Chromosome 23 haplotypes (blue) are inherited IBD in both affected dogs.
Sequencing of CHRNG, CHRND, and COLQ
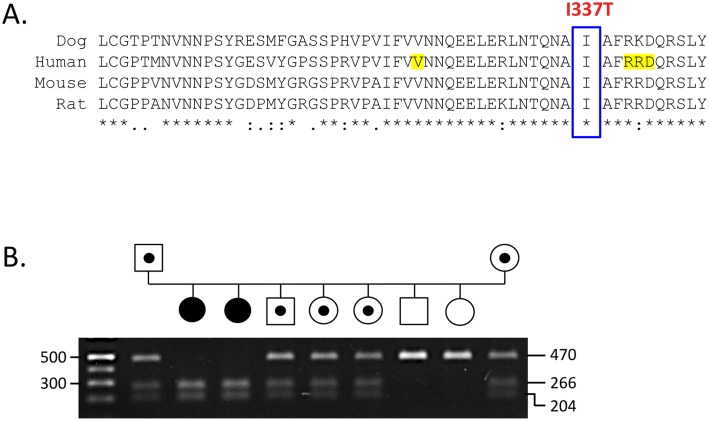
No nonsynonymous variants were identified in CHRNG. A single nonsynonymous variant in exon 4 of CHRND did not segregate with a recessive phenotype. Sequence data for COLQ revealed 3 nonsynonymous variants, only 1 of which segregated with a recessive trait. The exon 14 variant, c.1010T>C, predicts the substitution of isoleucine with threonine at residue 337 (Figure 6A).
Figure 6. (A) Sequence from the 5′ end of the C-terminal domain of ColQ in mammals.
Identical residues are denoted by an asterisk, conserved substitutions by a colon, and semi-conserved substitutions by a period. Residues altered in human CMS cases are highlighted in yellow [4], [30], [32], [37]. (B) BtsI digest results for the Labrador Retriever family. PCR amplicons from COLQ exon 14 are 470 bp in size and cleaved into 204 and 266 bp fragments in the presence of c.1010T>C. Three clinically normal littermates were identified as carriers, denoted by semi-filled icons.
Screening of c.1010T>C Variant
PCR amplicons from the COLQ exon 14 primer set are 470 bp in size. BtsI cleaves the amplicon only in individuals having the c.1010T>C allele, yielding fragments of 204 bp and 266 bp. Heterozygotes for the variant have all 3 fragments sizes (Figure 6B).
Digestion with Bts1 was used to genotype the variant for 49 additional members of this Labrador Retriever family, 288 unrelated Labrador Retrievers, and 112 dogs representing 65 other breeds (Table S3). Of the 58 family members, 40 were homozygous wild-type, 16 were heterozygous, and only the 2 affected dogs described herein were homozygous for the variant. The variant was not present in any unrelated Labrador Retrievers or dogs from other breeds.
Discussion
The probands in this study presented with an early onset neuromuscular disorder characterized by severe exercise-induced weakness. The lack of specific morphological changes in muscle and peripheral nerve biopsies excluded an underlying congenital myopathy or neuropathy. Electrodiagnostic findings and decreased AChR concentration in the muscle indicated a disorder of neuromuscular transmission. The autoimmune disease myasthenia gravis was eliminated based on the early age of onset and an absence of AChR antibodies in serum and AChR-bound antibodies in the muscle. The clinical diagnosis in the Labrador Retrievers was CMS.
While clinical signs and electrophysiological findings are generally similar between presynaptic, synaptic, and postsynaptic forms of CMS, a notable observation in the affected puppies was a worsening of the phenotype upon administration of an AChE inhibitor. This response indicates desensitization of the AChRs from overexposure to ACh and is consistent with a synaptic form of CMS referred to as end-plate AChE deficiency (EAD) [5]. EAD accounts for 10% to 15% of all human cases of CMSs and is always caused by mutations in COLQ [5]. COLQ encodes a collagen strand that homotrimerizes to form the tail subunit of asymmetric AChE. ColQ anchors AChE to the basal lamina where the enzyme hydrolyzes ACh, thereby limiting the length of the synaptic response [28]. In the absence of ColQ, ACh accumulates, causing prolonged muscle contraction and eventually the desensitization of AChR [29].
Through the examination of SNP allele frequencies in the Labrador Retriever family, we identified 2 chromosomes harboring CMS candidate genes that showed an inheritance pattern consistent with autosomal recessive transmission. Whereas human forms of CMS are often caused by compound heterozygosity, low levels of genetic diversity within purebred dog populations make simple recessive alleles more common. Linebreeding in this Labrador Retriever family makes it likely that the sire and dam inherited the mutation from a common ancestor and that the affected puppies are homozygous for a chromosome segment transmitted IBD. Analysis of polymorphic microsatellites showed that the regions flanking COLQ are IBD, whereas those flanking the other 2 identified candidate genes are not.
Sequencing of COLQ in the Labrador Retrievers revealed a missense mutation that predicts the replacement of a conserved hydrophobic isoleucine with a hydrophilic threonine in the C-terminal domain. ColQ has 3 domains: an N-terminal proline-rich attachment domain (PRAD), a collagenic central domain, and a C-terminal domain. The PRAD serves to attach the ColQ strand to an AChE tetramer. The collagen domain assembles the triple helix, while the C-terminal domain is involved in both the formation of the triple helix [30] and anchoring of the structure to the basal lamina [30], [31].
In humans, mutations responsible for EAD have been identified in each domain of COLQ and have different functional consequences depending on their location [4]. In the C-terminus, missense mutations in residues ranging from positions 342 to 452 are thought to inhibit the attachment of ColQ to the basal lamina of the muscle cell [30]–[33]. Some C-terminus mutations (e.g., V322D) may prevent the formation of the ColQ triple helix [32]. In the affected Labrador Retrievers, localization of the esterase reaction showed a poor correlation between AChE and AChR. This finding suggests improper anchoring of ColQ to the basal lamina, or mislocalization. Insufficient muscle samples prevented us from conducting a sedimentation profile of AChE to determine the exact consequence of the I337T mutation identified.
Linebreeding practices expedite the appearance of recessive diseases in purebred dog populations. The availability of genetic tests for the detection of carrier dogs allows for selective breeding to prevent widespread dissemination of the deleterious allele to the breed while maintaining genetic diversity. Because only 2 affected littermates were available for study herein, GWAS techniques could not be applied. The analysis of chromosomal inheritance patterns indicated a single functional and positional candidate gene and led to the discovery of the COLQ c.1010T>C mutation; however, our approach does not exclude the possibility that another mutation exists in a novel CMS gene.
While this manuscript was in revision, Matlik et al. reported that an identical mutation (c.1010T>C; I337T) was homozygous in 2 human CMS patients with EAD [34]. The affected children were first cousins from consanguineous relationships; both sets of parents were heterozygous for the mutation [34]. The substitution was the only variation identified in COLQ and was determined to be pathogenic through a prediction program [34]. Although uncommon, identical changes at the DNA level between humans and dogs with similar phenotypes have been previously identified [35], [36]. The identification of c.1010T>C in humans and dogs diagnosed with CMS strongly supports the causality of the mutation and shows that conservation of residue 337 is critical for proper function of ColQ.
Supporting Information
Primers (5′-3′) for amplification of CHRNG, CHRND, and COLQ. Primers were designed to amplify exons and splice sites. ExoSAP indicates the use of the ExoSAP protocol and Gel X indicates the use of the gel extraction protocol for post-PCR clean-up.
(PDF)
Candidate regions based on allele frequencies. Genomic regions known to harbor CMS candidate genes were screened for case allele frequencies of 1.0 and control allele frequencies of between 0.14 and 0.50. CHR = chromosome number; SNP = SNP name; BP = chromosome position; A1 = allele 1; F_A = frequency of allele 1 in affected dogs (cases); F_U = frequency of allele 1 in unaffected dogs (controls); A2 = allele 2.
(PDF)
Dogs screened for the COLQ 14 variant. Digestion with Bts1 was used to genotype the 2 affected dogs, 56 other members of the Labrador Retriever pedigree, 288 unrelated Labrador Retrievers, and 112 dogs representing 65 other breeds.
(PDF)
Acknowledgments
The authors would like to thank the dog owners and veterinarians who contributed DNA samples for this study.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R15AR062868 (LAC) and the Canine Health Foundation under Award Number 01311 (ANSM). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Canine Health Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hantaï D, Nicole S, Eymard B (2013) Congenital myasthenic syndromes: an update. Curr Opin Neurol 26: 561–8. [DOI] [PubMed] [Google Scholar]
- 2. Ohno K, Brengman J, Tsujino A, Engel AG (1998) Human endplate acetylcholinesterase deficiency caused by mutations in the collagen-like tail subunit (ColQ) of the asymmetric enzyme. Proc Natl Acad Sci USA 95: 9654–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ishigaki K, Nicolle D, Krejci E, Leroy J-P, Koenig J, et al. (2003) Two novel mutations in the COLQ gene cause endplate acetylcholinesterase deficiency. Neuromuscul Disord 13: 236–244. [DOI] [PubMed] [Google Scholar]
- 4. Mihaylova V, Müller JS, Vilchez JJ, Salih MA, Kabiraj MM, et al. (2008) Clinical and molecular genetic findings in COLQ-mutant congenital myasthenic syndromes. Brain 131: 747–759. [DOI] [PubMed] [Google Scholar]
- 5.Abicht A, Müller JS, Lochmüller H (2012) Congenital Myasthenic Syndromes. GeneReviews Available: http://www.ncbi.nlm.nih.gov/books/NBK1168/. Accessed 27 February 2014.
- 6. Palmer AC, Barker J (1974) Myasthenia in the dog. Vet Rec 95: 452–454. [DOI] [PubMed] [Google Scholar]
- 7. Oda K, Lambert EH, Lennon VA, Palmer AC (1984) Congenital canine myasthenia gravis: I. Deficient junctional acetylcholine receptors. Muscle Nerve 7: 705–716. [DOI] [PubMed] [Google Scholar]
- 8. Miller LM, Lennon VA, Lambert EH, Reed SM, Hegreberg GA, et al. (1983) Congenital myasthenia in 13 smooth fox terriers. J Am Vet Med Assoc 182: 694–697. [PubMed] [Google Scholar]
- 9. Johnson RP, Watson AD, Smith J, Cooper BJ (1975) Myasthenia in Springer Spaniel littermates. J Small Anim Pract 16: 641–647. [DOI] [PubMed] [Google Scholar]
- 10. Dickinson PJ, Sturges BK, Shelton GD, LeCouteur RA (2005) Congenital Myasthenia Gravis in Smooth-Haired Miniature Dachshund Dogs. J Vet Intern Med 19: 920–923. [DOI] [PubMed] [Google Scholar]
- 11. Flagstad A, Trojaborg W, Gammeltoft S (1989) Congenital myasthenic syndrome in the dog breed Gammel Dansk Hønsehund: clinical, electrophysiological, pharmacological and immunological comparison with acquired myasthenia gravis. Acta Vet Scand 30: 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Proschowsky HF, Flagstad A, Cirera S, Joergensen CB, Fredholm M (2007) Identification of a Mutation in the CHAT Gene of Old Danish Pointing Dogs Affected with Congenital Myasthenic Syndrome. J Hered 98: 539–543. [DOI] [PubMed] [Google Scholar]
- 13. Kraner S, Sieb JP, Thompson PN, Steinlein OK (2002) Congenital myasthenia in Brahman calves caused by homozygosity for a CHRNE truncating mutation. Neurogenetics 4: 87–91. [DOI] [PubMed] [Google Scholar]
- 14. Patterson EE, Minor KM, Tchernatynskaia AV, Taylor SM, Shelton GD, et al. (2008) A canine DNM1 mutation is highly associated with the syndrome of exercised-induced collapse. Nat Genet 40: 1235–1239. [DOI] [PubMed] [Google Scholar]
- 15. Pelé M, Tiret L, Kessler J-L, Blot S, Panthier J-J (2005) SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet 14: 1417–1427. [DOI] [PubMed] [Google Scholar]
- 16. Beggs AH, Bohm J, Snead E, Kozlowski M, Maurer M, et al. (2010) MTM1 mutation associated with X-linked myotubular myopathy in Labrador retrievers. Proc Natl Acad Sci U S A 107: 14697–14702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gill JL, Tsai KL, Krey C, Noorai RE, Vanbellinghen J-F, et al. (2012) A canine BCAN microdeletion associated with episodic falling syndrome. Neurobiol Dis 45: 130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Safra N, Bassuk AG, Ferguson PJ, Aguilar M, Coulson RL, et al. (2013) Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, NKX2–8, as a Genetic Component of Neural Tube Defects in Humans. PLoS Genet 9: e1003646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vernau KM, Runstadler JA, Brown EA, Cameron JM, Huson HJ, et al. (2013) Genome-Wide Association Analysis Identifies a Mutation in the Thiamine Transporter 2 (SLC19A3) Gene Associated with Alaskan Husky Encephalopathy. PLoS One 8: e57195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubowitz V, Sewry CA (2007). Histological and histochemical stains and reactions. In: Dubowitz V, Sewry CA, editors. Muscle Biopsy: A Practical Approach, 3rd ed. London: Saunders Elsevier: pp. 21–39.
- 21. Lindstrom JM, Lambert EH (1978) Content of acetylcholine receptor and antibodies bound to receptor in myasthenia gravis, experimental autoimmune myasthenia gravis, and Eaton-Lambert syndrome. Neurology 28: 130–138. [DOI] [PubMed] [Google Scholar]
- 22. Lindstrom J, Shelton D, Fujii Y (1988) Myasthenia gravis. Adv Immunol 42: 233–284. [DOI] [PubMed] [Google Scholar]
- 23. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cargill EJ, Clark LA, Steiner JM, Murphy KE (2002) Multiplexing of canine microsatellite markers for whole-genome screens. Genomics 80: 250–3. [DOI] [PubMed] [Google Scholar]
- 25. Clark LA, Tsai KL, Steiner JM, Williams DA, Guerra T, et al. (2004) Chromosome-specific microsatellite multiplex sets for linkage studies in the domestic dog. Genomics 84: 550–554. [DOI] [PubMed] [Google Scholar]
- 26. Swallow JS, Griffiths IR (1977) Age related changes in the motor nerve conduction velocity in dogs. Res Vet Sci 23: 29–32. [PubMed] [Google Scholar]
- 27. Sims MH, Redding RW (1980) Maturation of nerve conduction velocity and the evoked muscle potential in the dog. Am J Vet Res 41: 1247–52. [PubMed] [Google Scholar]
- 28. Katz B, Miledi R (1973) The Binding of Acetylcholine to Receptors and its Removal from the Synaptic Cleft. J Physiol 231: 549–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Engel AG, Lambert EH, Gomez MR (1977) A New Myasthenic Syndrome with End-Plate Acetylcholinesterase Deficiency, Small Nerve Terminals, and Reduced Acetylcholine Release. Ann Neurol 1: 315–330. [DOI] [PubMed] [Google Scholar]
- 30. Ohno K, Engel AG, Brengman JM, Shen X-M, Heidenreich F, et al. (2000) The Spectrum of Mutations Causing End-Plate Acetylcholinesterase Deficiency. Ann Neurol 47: 162–170. [PubMed] [Google Scholar]
- 31. Kimbell LM, Ohno K, Engel AG, Rotundo RL (2004) C-terminal and Heparin-binding Domains of Collagenic Tail Subunit Are Both Essential for Anchoring Acetylcholinesterase at the Synapse. J Biol Chem 279: 10997–11005. [DOI] [PubMed] [Google Scholar]
- 32. Nakata T, Ito M, Azuma Y, Otsuka K, Noguchi Y, et al. (2013) Mutations in the C-Terminal Domain of ColQ in Endplate Acetylcholinesterase Deficiency Compromise ColQ–MuSK Interaction. Hum Mutat 00: 1–8. [DOI] [PubMed] [Google Scholar]
- 33. Arredondo J, Lara M, Ng F, Gochez DA, Lee DC, et al. (2014) COOHterminal collagen Q (COLQ) mutants causing human deficiency of endplate acetylcholinesterase impair the interaction of ColQ with proteins of the basal lamina. Hum Genet 133: 599–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matlik HN, Milhem RM, Saadeldin IY, Al-Jaibeji HS, Al-Gazali L, et al. (2014) Clinical and Molecular Analysis of a Novel COLQ Missense Mutation Causing Congenital Myasthenic Syndrome in a Syrian Family. Pediatr Neurol 51: 165–169. [DOI] [PubMed] [Google Scholar]
- 35. Zangerl B, Goldstein O, Philp AR, Lindauer SJ, Pearce-Kelling SE, et al. (2006) Identical mutation in a novel retinal gene causes progressive rod-cone degeneration in dogs and retinitis pigmentosa in humans. Genomics 88: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seppälä EH, Reuser AJ, Lohi H (2013) A nonsense mutation in the acid α-glucosidase gene causes Pompe disease in Finnish and Swedish Lapphunds. PLoS One 8: e56825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wargon I, Richard P, Kuntzer T, Sternberg D, Nafissi S, et al. (2012) Long-term follow-up of patients with congenital myasthenic syndrome caused by COLQ mutations. Neuromuscul Disord 22: 318–324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers (5′-3′) for amplification of CHRNG, CHRND, and COLQ. Primers were designed to amplify exons and splice sites. ExoSAP indicates the use of the ExoSAP protocol and Gel X indicates the use of the gel extraction protocol for post-PCR clean-up.
(PDF)
Candidate regions based on allele frequencies. Genomic regions known to harbor CMS candidate genes were screened for case allele frequencies of 1.0 and control allele frequencies of between 0.14 and 0.50. CHR = chromosome number; SNP = SNP name; BP = chromosome position; A1 = allele 1; F_A = frequency of allele 1 in affected dogs (cases); F_U = frequency of allele 1 in unaffected dogs (controls); A2 = allele 2.
(PDF)
Dogs screened for the COLQ 14 variant. Digestion with Bts1 was used to genotype the 2 affected dogs, 56 other members of the Labrador Retriever pedigree, 288 unrelated Labrador Retrievers, and 112 dogs representing 65 other breeds.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.