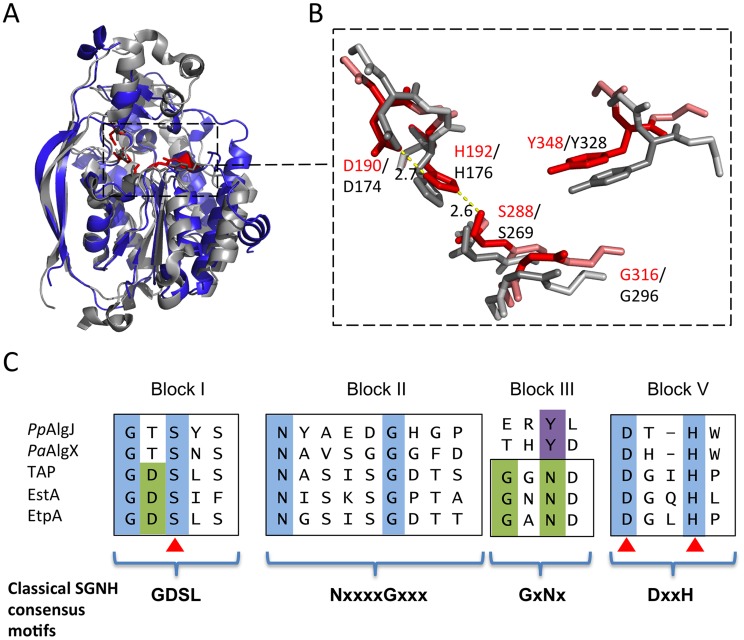
Figure 3. Architecture of the proposed PpAlgJ75–370 active site compared to AlgX.
(A) Superposition of the cartoon representations of PpAlgJ75–370 (blue) and PaAlgX44–344 (grey). The conserved residues implicated in catalysis in SGNH hydrolases are displayed as sticks and coloured either red (PpAlgJ) or grey (AlgX). The dashed box indicates the region where the conserved residues are located. (B) Enlarged figure highlighting the similarity of the residues in the active site region in both proteins. Red stick residues and text identifies the conserved residues from PpAlgJ whereas the grey sticks and black text identifies residues from AlgX. Yellow dashed lines represent hydrogen bonding between members of the catalytic triad with values in angstroms (Å). (C) Sequence alignment of various members of the SGNH hydrolase superfamily depicting four consensus blocks shared by members. PpAlgJ: P. aeruginosa O-acetyltransferase (Q88ND3), PaAlgX: P. aeruginosa O-acetyltransferase (Q51372) EstA: Lactobacillus helveticus arylesterase (Q9LAH7), EtpA: Vibrio mimicus arylesterase (Q07792). Conserved residues are masked in blue (absolutely conserved) or green (conserved in most members). Residues masked in purple correspond to the conserved tyrosine in AlgJ and AlgX that are proposed to be analogous in function to asparagine in other superfamily members. Brackets correspond to Uniprot identifiers.