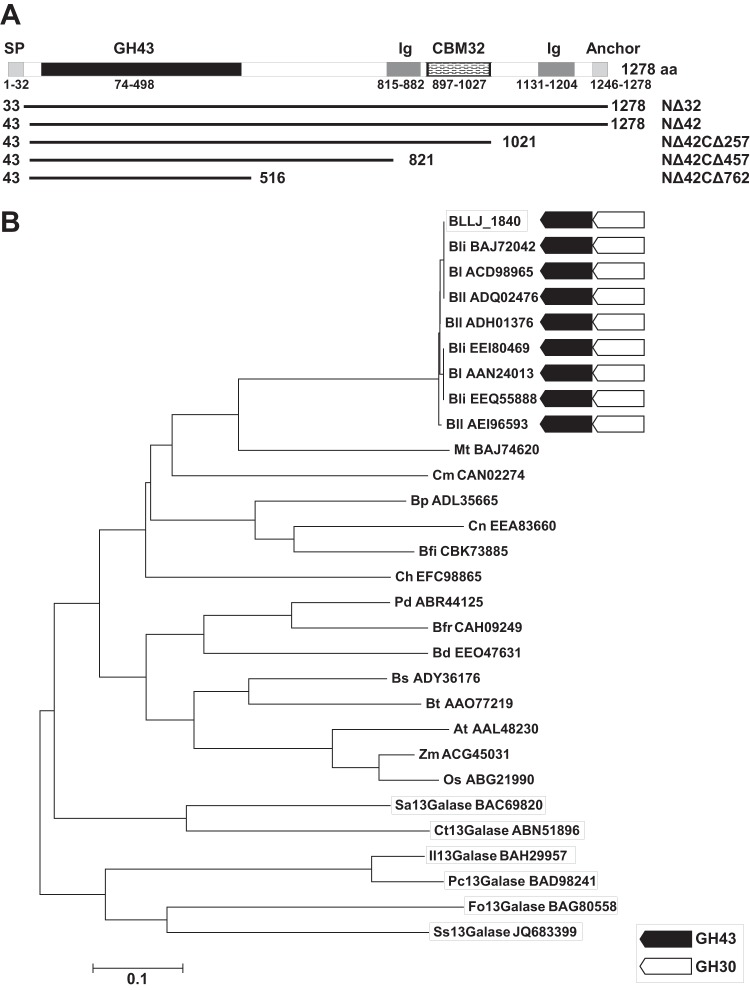
FIG 1.
Domain structures and phylogenetic relationships of BLLJ_1840. (A) BLLJ_1840 comprises the GH43 catalytic domain, bacterial Ig-like domain (Ig), CBM32, and the LPXTG cell wall anchor domain (Anchor). The signal peptide (SP) was predicted using the SignalP (version 4.1) server (http://www.cbs.dtu.dk/services/SignalP/). The lines indicate the length of the BLLJ_1840 deletion mutants. (B) The phylogenetic tree of BLLJ_1840 with homologous proteins was constructed with the neighbor-joining method using the aligned sequences with the MUSCLE program implemented in MEGA5 software. BLLJ_1840 and the characterized exo-β-1,3-galactanases are boxed. Black and white arrows, conservation of GH43 exo-β-1,3-galactanases and the neighboring GH30 endo-β-1,6-galactanase candidates, respectively. The GenBank accession numbers are shown with the abbreviations for the organisms, which are as follows: Bli, B. longum subsp. infantis; Bl, B. longum; Bll, B. longum subsp. longum; Mt, Microbacterium testaceum; Cm, Clavibacter michiganensis; Bp, Butyrivibrio proteoclasticus; Cn, Clostridium nexile; Bfi, Butyrivibrio fibrisolvens; Ch, Clostridium hathewayi; Pd, Parabacteroides distasonis; Bfr, Bacteroides fragilis; Bd, Bacteroides dorei; Bs, Bacteroides salanitronis; Bt, Bacteroides thetaiotaomicron; At, Arabidopsis thaliana; Zm, Zea mays; Os, Oryza sativa; Sa, Streptomyces avermitilis; Ct, Clostridium thermocellum; Il, Irpex lacteus; Pc, Phanerochaete chrysosporium; Fo, Fusarium oxysporum; Ss, Streptomyces sp.