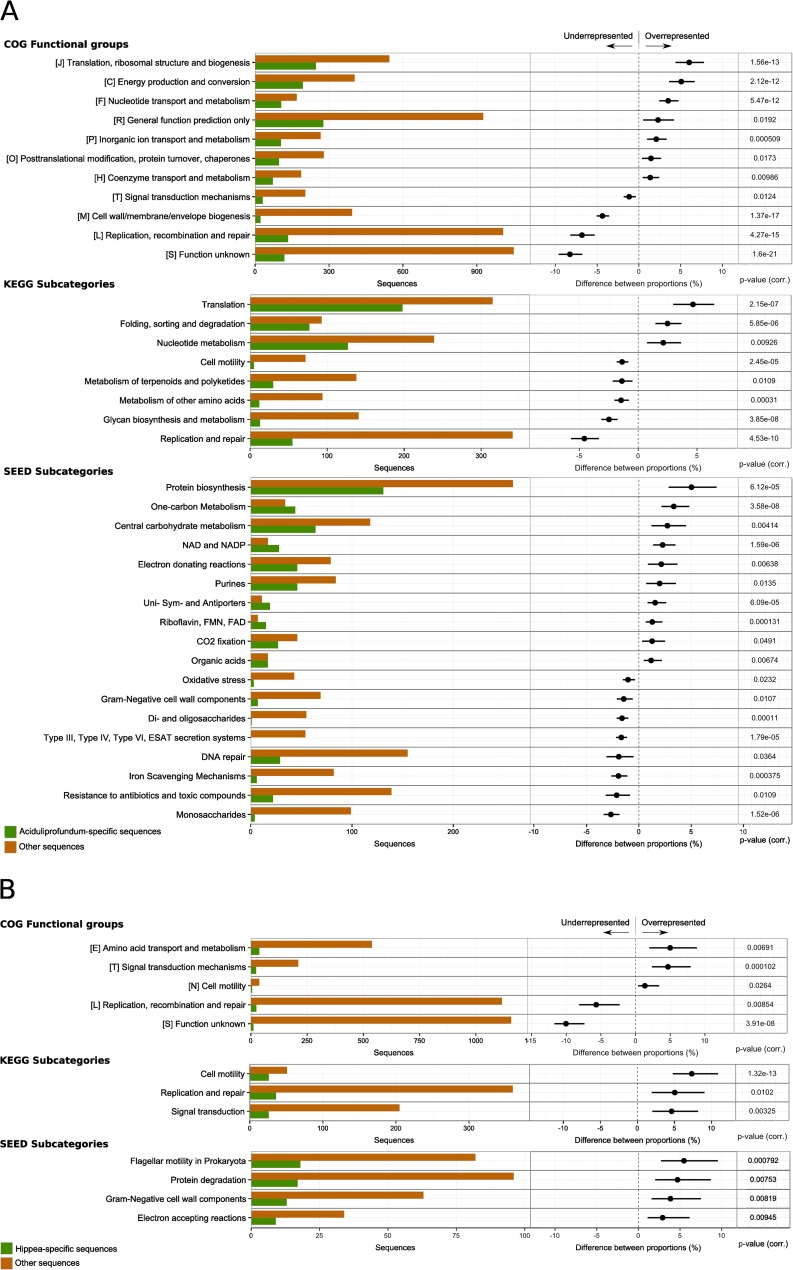
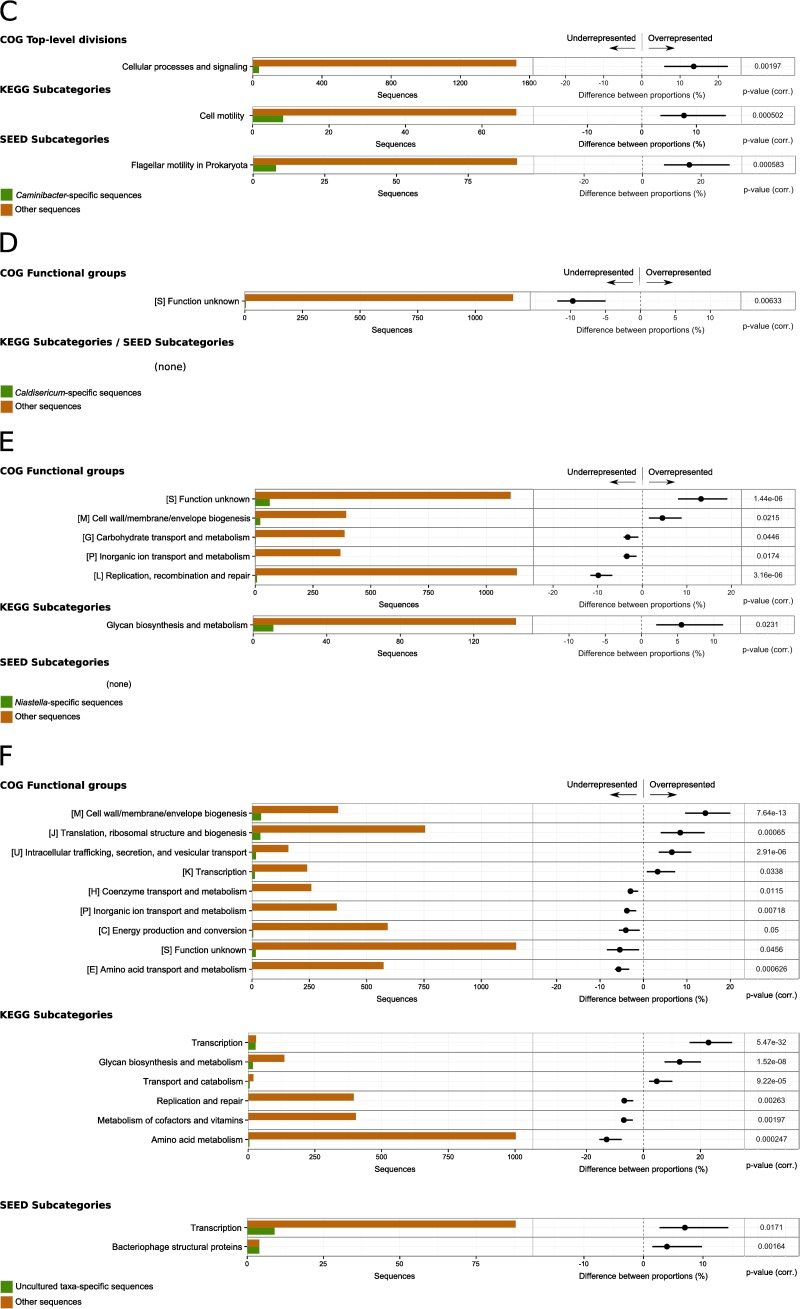
FIG 5.
Differential analysis of the functional assignments for subsets of the genes assigned to taxonomic groups, compared to the rest of the SP data set. The analyses results are shown for the five most abundant genera—Aciduliprofundum (A), Hippea (B), Caminibacter (C), Caldisericum (D), and Niastella (E)—and for uncultured taxa (F). Each row shows absolute counts (left) and differences between relative counts (right) for a functional category. In particular, the histograms on the left report the total number of genes assigned to the functional category in the taxonomic group of interest (green) and in the rest of the data set (orange). The dots in the bar plots on the right represent the difference (taxonomic group of interest minus the rest of the data set) between the proportions of genes assigned to the functional category, relative to the total counts in the subset. The extent of the bar represents the 95% confidence intervals of the difference between proportions. Only data with a corrected P value of ≤0.05 (reported on the right) and a difference between proportions of ≥1.0% are reported.