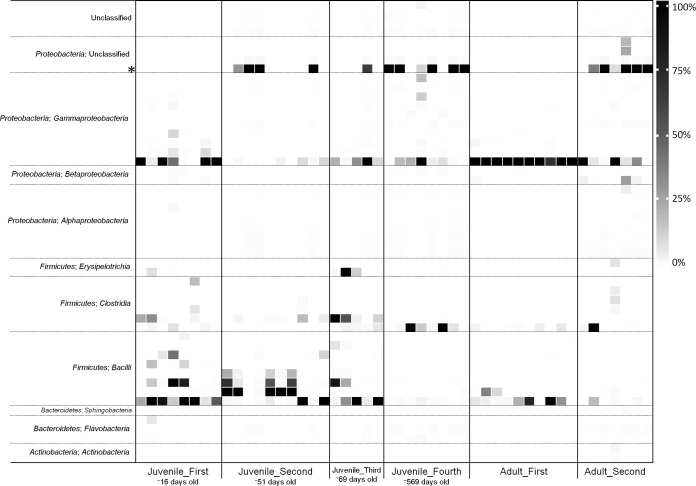
FIG 1.
Phylogenetic distribution of bacterial OTUs in the kakapo fecal microbiota. High-level taxonomic information is provided as per classification. OTUs are defined as groups of 16S rRNA gene sequences that share >97% similarity and are ordered by phylum and then subordered by class. For clarity, only the 50 most abundant OTUs are plotted, representing 98.0% of the total reads, with 477 OTUs comprising the remainder of the reads following removal of singletons. OTU abundances are scaled as a proportion of all sequences in the respective sample. OTU02 (mentioned in the text) is noted with an asterisk.